Chimera Workshop
November 17-18, 2005
Demonstration of Chimera multiscale tool for:

| 
|
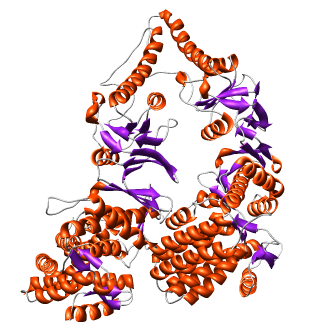
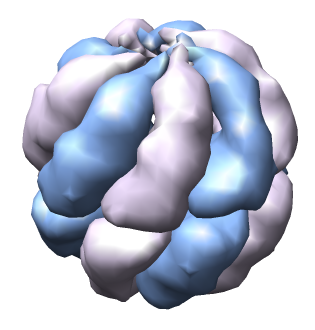
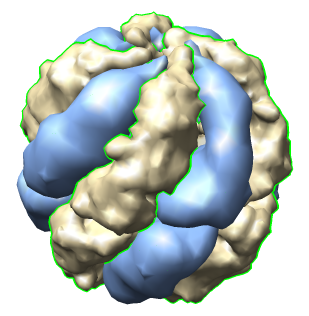
| Thermosome model 1A6D has asymmetric unit consisting of 2 proteins with 61% sequence identity. | Biological unit made with Multiscale tool is 8 copies of asymmetric unit. |
The 1A6D PDB file contains 8 matrices in REMARK 350 that define the biological unit and were read by Chimera.
REMARK 350 GENERATING THE BIOMOLECULE
REMARK 350 COORDINATES FOR A COMPLETE MULTIMER REPRESENTING THE KNOWN
REMARK 350 BIOLOGICALLY SIGNIFICANT OLIGOMERIZATION STATE OF THE
REMARK 350 MOLECULE CAN BE GENERATED BY APPLYING BIOMT TRANSFORMATIONS
REMARK 350 GIVEN BELOW. BOTH NON-CRYSTALLOGRAPHIC AND
REMARK 350 CRYSTALLOGRAPHIC OPERATIONS ARE GIVEN.
REMARK 350
REMARK 350 APPLY THE FOLLOWING TO CHAINS: A, B
REMARK 350 BIOMT1 1 1.000000 0.000000 0.000000 0.00000
REMARK 350 BIOMT2 1 0.000000 1.000000 0.000000 0.00000
REMARK 350 BIOMT3 1 0.000000 0.000000 1.000000 0.00000
REMARK 350 BIOMT1 2 -1.000000 0.000000 0.000000 0.00000
REMARK 350 BIOMT2 2 0.000000 -1.000000 0.000000 0.00000
REMARK 350 BIOMT3 2 0.000000 0.000000 1.000000 0.00000
REMARK 350 BIOMT1 3 0.000000 -1.000000 0.000000 0.00000
REMARK 350 BIOMT2 3 1.000000 0.000000 0.000000 0.00000
REMARK 350 BIOMT3 3 0.000000 0.000000 1.000000 0.00000
REMARK 350 BIOMT1 4 0.000000 1.000000 0.000000 0.00000
REMARK 350 BIOMT2 4 -1.000000 0.000000 0.000000 0.00000
REMARK 350 BIOMT3 4 0.000000 0.000000 1.000000 0.00000
REMARK 350 BIOMT1 5 -1.000000 0.000000 0.000000 0.00000
REMARK 350 BIOMT2 5 0.000000 1.000000 0.000000 0.00000
REMARK 350 BIOMT3 5 0.000000 0.000000 -1.000000 0.00000
REMARK 350 BIOMT1 6 1.000000 0.000000 0.000000 0.00000
REMARK 350 BIOMT2 6 0.000000 -1.000000 0.000000 0.00000
REMARK 350 BIOMT3 6 0.000000 0.000000 -1.000000 0.00000
REMARK 350 BIOMT1 7 0.000000 1.000000 0.000000 0.00000
REMARK 350 BIOMT2 7 1.000000 0.000000 0.000000 0.00000
REMARK 350 BIOMT3 7 0.000000 0.000000 -1.000000 0.00000
REMARK 350 BIOMT1 8 0.000000 -1.000000 0.000000 0.00000
REMARK 350 BIOMT2 8 -1.000000 0.000000 0.000000 0.00000
REMARK 350 BIOMT3 8 0.000000 0.000000 -1.000000 0.00000
| 
|
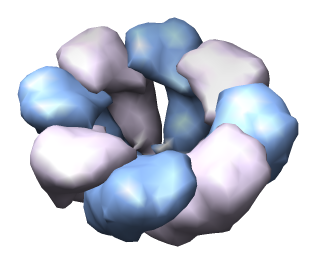
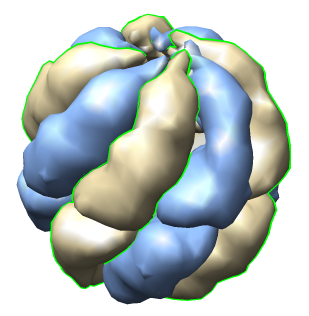
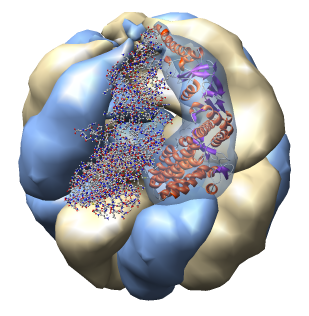
| Half of shell hidden. | Select copies of one protein and recolor. |
| 
|
| Change resolution of surfaces 8A -> 4A. | Show ribbon and ball and stick. Add transparent surface. |
| |
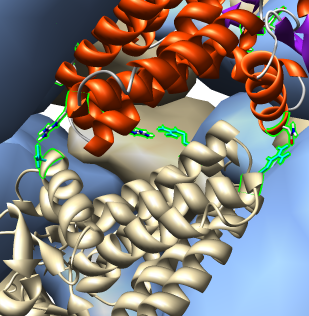
| Show contacts within 2A of chain A with "load atoms" switch on. Show when extra copies are created in Model Panel. Promote contact atoms to residues, show as stick, and identify contact residues by hovering mouse over them. |