Relational Database Technology
John "Scooter" Morris
April 6, 2017
Overview
- Data modeling review
- Relational algebra
- SQL
- From model to schema
Limitations
- What am I not telling you about?
- database normalization
- object-based approaches to database design
- object-relational mapping
- .... too much more to mention ....
- Ask questions!
Example Problem
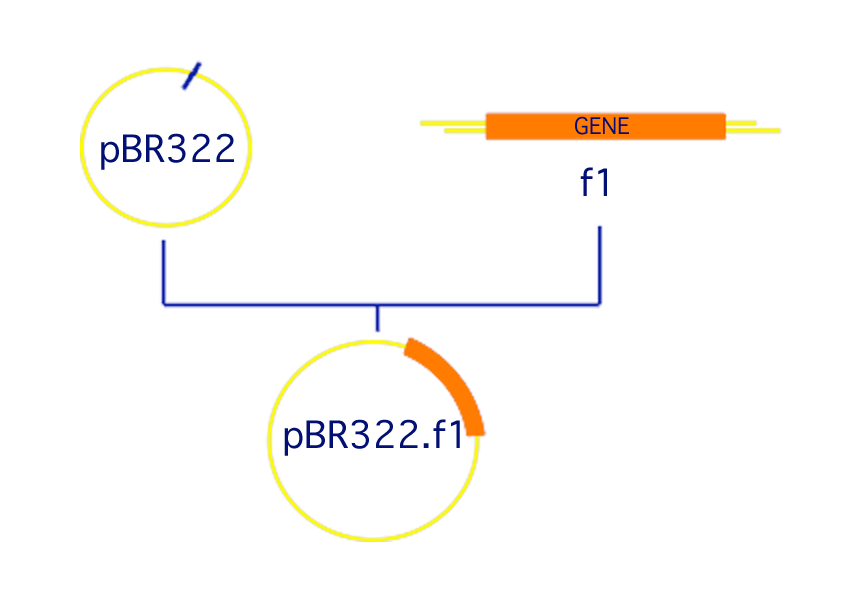
A system to automate the tracking and documentation of plasmid construction- Terminology:
- fragment: a length of double-stranded DNA
- plasmid: a circular fragment
- recipe: a series of manipulations of the DNA to produce a new plasmid with cDNA of interest inserted
- ACL: access control list
- Needs:
- Data processing -- convert raw data into results
- Visualization -- a way to visualize the results
- Data storage -- store the results (and perhaps the raw data)
Example problem
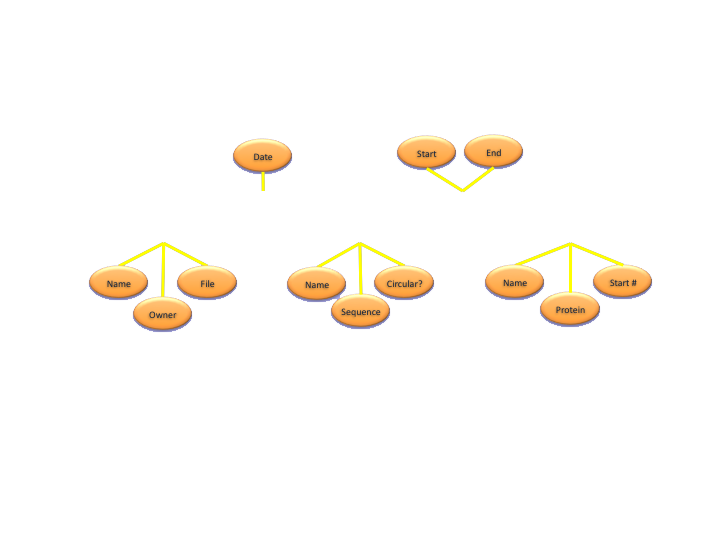
Data Modeling
- The FIRST Step
- Structured way to understand the data semantics
- Independent of underlying platform
- Way to communicate with team members (including users)
- Excellent (minimal?) documentation
- Example: ER Diagrams
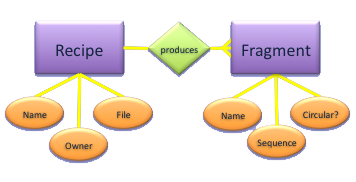
ER Diagrams


Types of Databases
- Flat-file
- Hierarchical
- Network
- Relational
- Object
- Object-Relational
Flat-File Databases
- No database-enforced (or provided) linkage between records
- Excellent for small or special-purpose databases
- Might include support for single or multiple indexes
- Major feature: ease of use (Filemaker, Access)
- Major drawback: scalability & flexibility
- e.g.:
- ndbm
- Berkeley DB (Sleepycat DB)
- vi, grep, sed
- FileMaker
- Access
Hierarchical Databases
- Relationship between Recipe and Fragment is one-to-many (master-detail)
- Assume two recipes: r1.cr and r2.cr
- r1.cr produces 2 plasmids and 1 fragment:
- r1.p1, r1.p2, and r1.f1
- r2.cr produces 2 fragments:
- r2.f1, and r2.f2
- r1.cr produces 2 plasmids and 1 fragment:
Hierarchical Databases
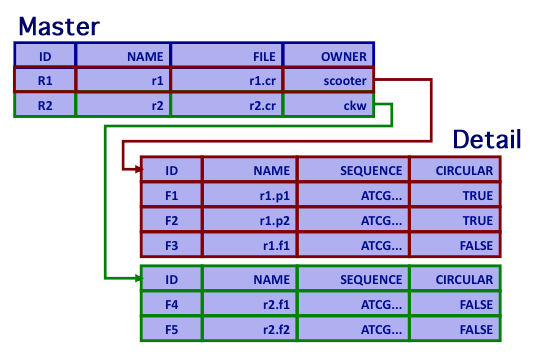
Hierarchical Databases
- Database provides explicit master-detail support
- Ideal for many business applications
- Restricted to a strict hierarchy
- Queries down the hierarchy are very efficient
- Any other queries are very expensive
- e.g.
- IMS
- What about many-to-many relationships?
Networked Databases
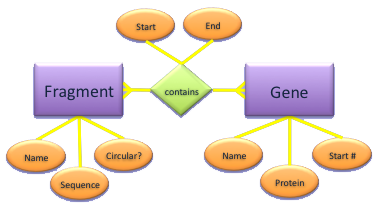
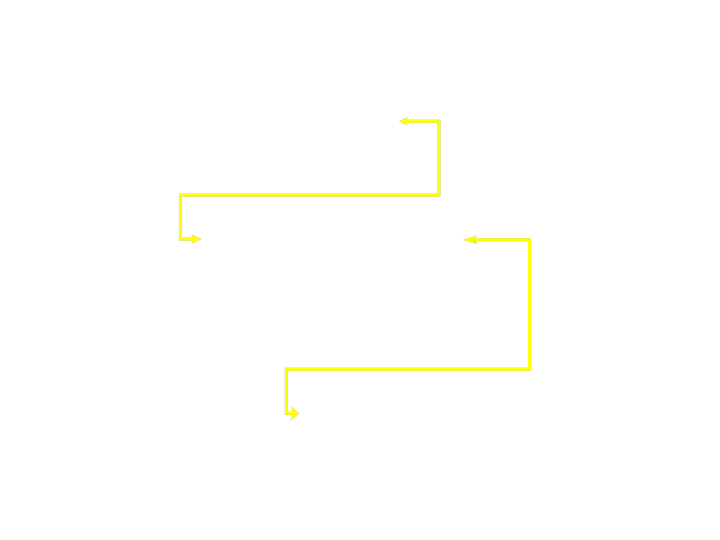
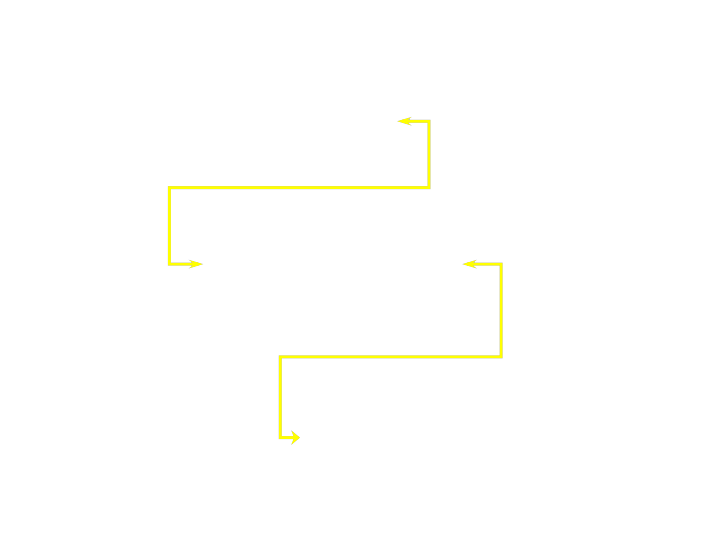
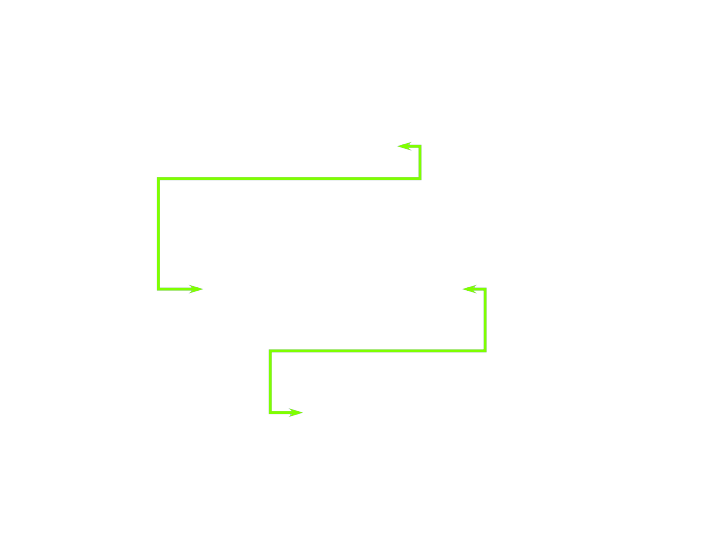
- Fragment and Gene have a many-to-many relationship
- Not represented well by hierarchical databases
Networked Databases
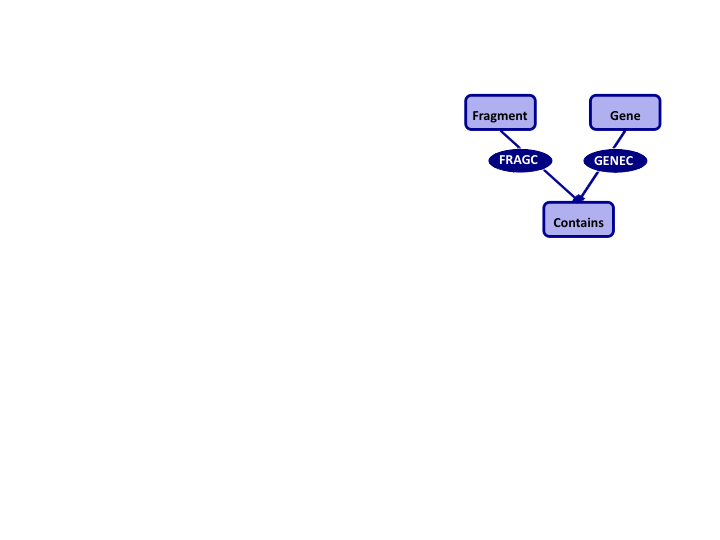

Networked Databases
- Based on set theory
- Database provides explicit linkage support
- Very significant design costs
- Queries along the connection path are very efficient
- Any other queries are very expensive
- e.g.
- CODASYL
- What if I want to "discover" other relationships?
Relational Databases
- Foundation of most production databases
- Based on relational calculus and relational algebra
- Allows ad-hoc query capability across record types
- Supports a standard query language (SQL)
- Can support either hierarchical or network models
- Attributes are limited to basic types
- e.g.
- SQLite
- MySQL
- Derby
- Oracle
- DB2
Relational Databases
- Based on relational views (tables)
- Associations are based on data values, not expressed linkages
- All data is expressed in tables
- Terminology:
- Rows are called tuples
- Columns (attributes) are of a common domain (type)
ER → Relational Schema
- First, combine any entities with a one-to-one relationship
- Next define tables for our entities:
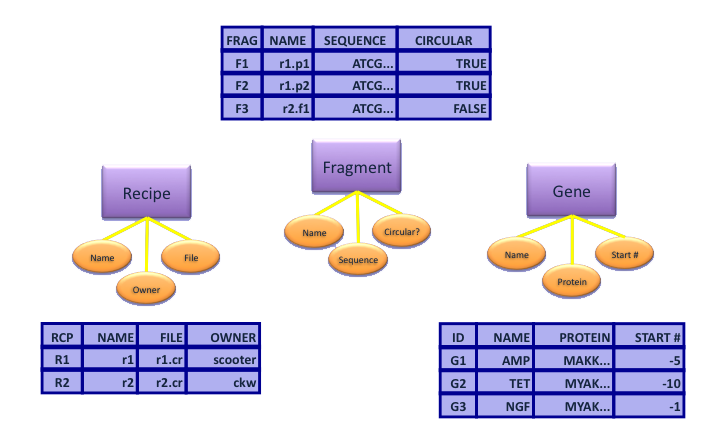
- Note that we've added a new attribute to serve as the primary key for each entity
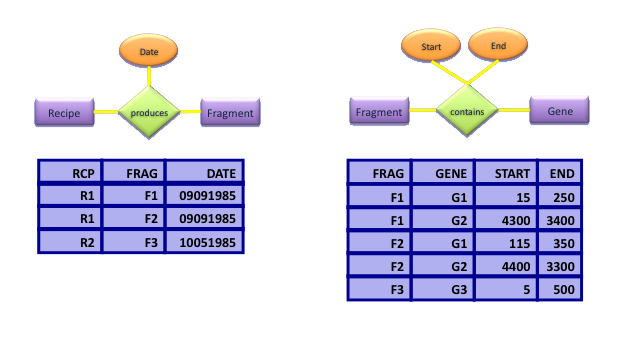
ER → Relational Schema
- Now define tables for relationships, adding attributes for the associations:
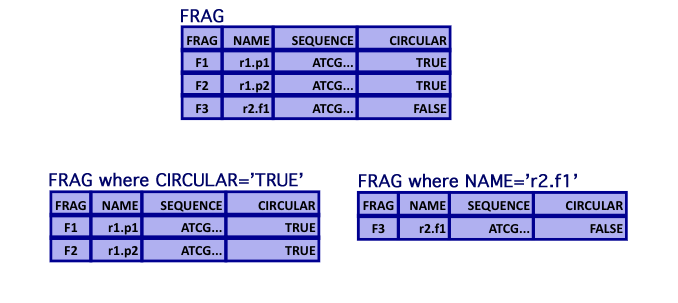
Relational Algebra: Selection
- Selection
- Selection of tuples based on Boolean criteria
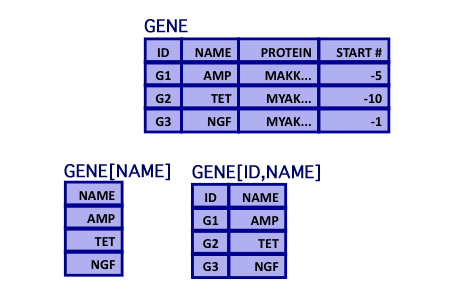
Relational Algebra: Projection
- Projection
- Selection of attributes
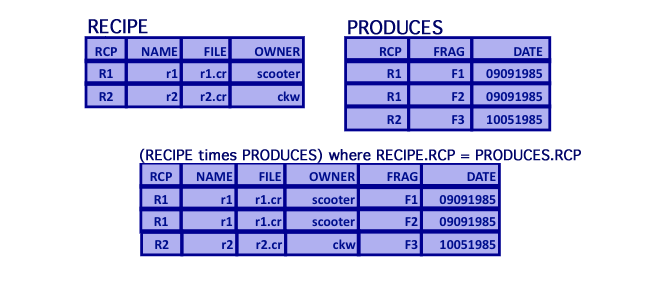
Relational Algebra: Inner Join
- Inner Join
- Matrix product of two relations based on a given join predicate, where each record in the two joined tables has a matching record.
- EquiJoin
- Inner join where the join predicate is based on an equality.
- Natural Join
- Inner join where the join predicate is implicitly based on attributes with the same name in each of the join tables
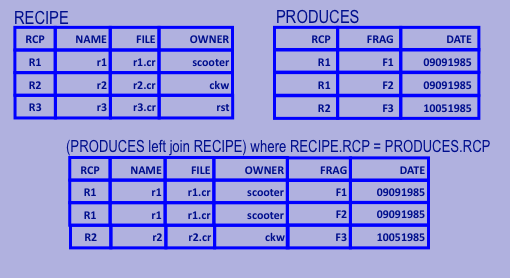
Relational Algebra: Outer Join
- Left Outer Join
- Join where the result contains all records from the left table, but not necessarily from the right.
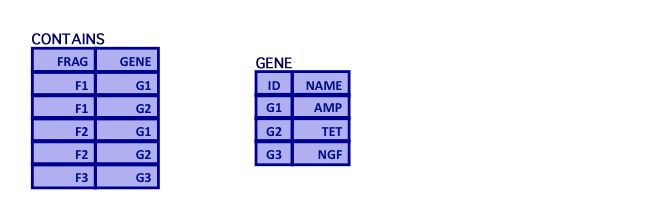
Relational Algebra: Example Query
- Query: What recipes produce the AMP gene?
- First, select the AMP gene from the GENE relation and join it to CONTAINS
TEMP1 = (CONTAINS[FRAG,GENE] times GENE[ID,NAME]) where GENE.ID=CONTAINS.GENE and GENE.NAME='AMP'
- First, select the AMP gene from the GENE relation and join it to CONTAINS
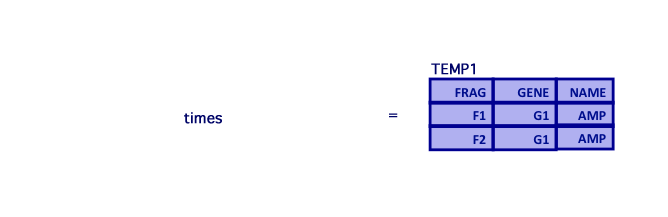
Relational Algebra: Example Query
- Query: What recipes produce the AMP gene?
- Second, join the result to the PRODUCES relation and select the RCP attribute
TEMP2 = (TEMP1 join PRODUCES)[RCP]*
- Second, join the result to the PRODUCES relation and select the RCP attribute
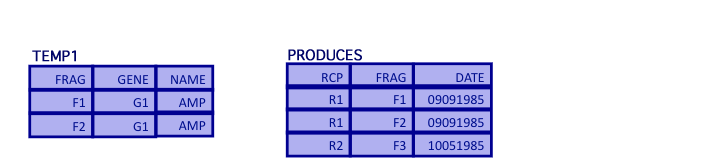

Relational Algebra: Example Query
- Query: What recipes produce the AMP gene?
- Finally, join the result to the RECIPES relation
ANSWER = (TEMP2 join RECIPES) where TEMP2.RCP=RECIPES.RCP
- Finally, join the result to the RECIPES relation
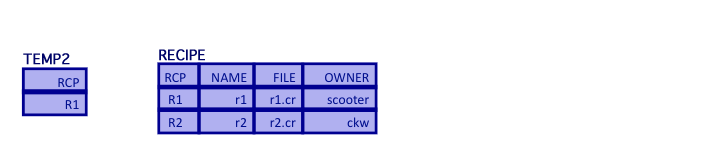
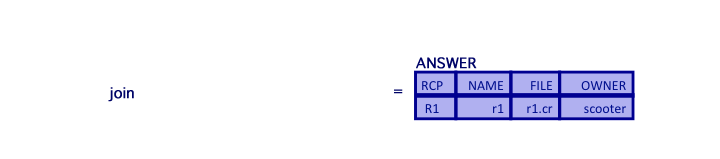
Structured Query Language (SQL)
- ANSI standard syntax for relational algebra
- Supported by all major commercial relational databases
- Also supported by many open-source efforts
- e.g. mysql, perl's DBI/DBD
- Will only cover:
- CREATE
- INSERT
- SELECT
- JOIN
- UPDATE
SQL - CREATE
- Creates database objects (databases, tables, indices)
- SYNOPSIS:
CREATE DATABASE database_nameCREATE TABLE table_name( column_name1 data_type, column_name2 data_type, ...... [PRIMARY KEY (column_name),] [FOREIGN KEY (column_name)REFERENCES table_name(column_name),]) CREATE [UNIQUE ]INDEX index_nameON table_name( column_name)
- SYNOPSIS:
SQL - CREATE
- Examples:
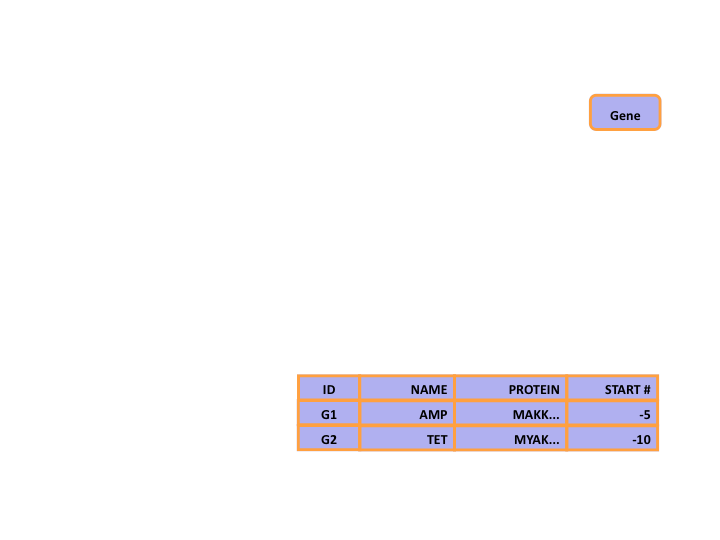
CREATE TABLE "GENE" ( ID char(16), NAME varchar(20), PROTEIN varchar, START int, PRIMARY KEY (ID) );CREATE TABLE "PRODUCES" ( RCP char(16), FRAG char(16), DATE date, FOREIGN KEY (RCP) REFERENCES RECIPE(RCP), FOREIGN KEY (FRAG) REFERENCES FRAG(ID) );CREATE UNIQUE INDEX on GENE (ID);
SQL - INSERT
- Inserts data into a table row
- SYNOPSIS:
INSERT INTO "tablename" (first_column,...last_column)VALUES (first_value,...last_value); - Example:
INSERT INTO GENE (ID, NAME, PROTEIN, START) VALUES ('G1', 'AMP', 'MAKK...', -5);
SQL - SELECT
- Selects data from relational tables
- Key syntax for expressing relational algebra
- SYNOPSIS
SELECT [DISTINCT ] column1[,column2]FROM table1[,table2] [WHERE "conditions"] [GROUP BY "column-list"] [HAVING "conditions] [ORDER BY "column-list" [ASC |DESC ] ]
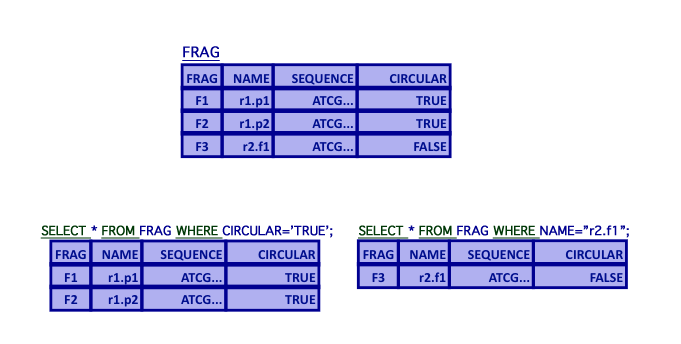
SELECT - Selection
- Selection
- Selection of tuples based on Boolean criteria
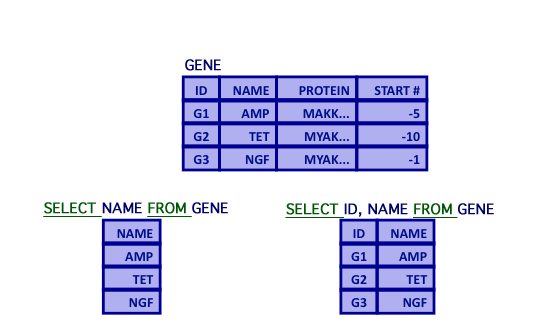
SELECT - Projection
- Projection
- Selection of attributes
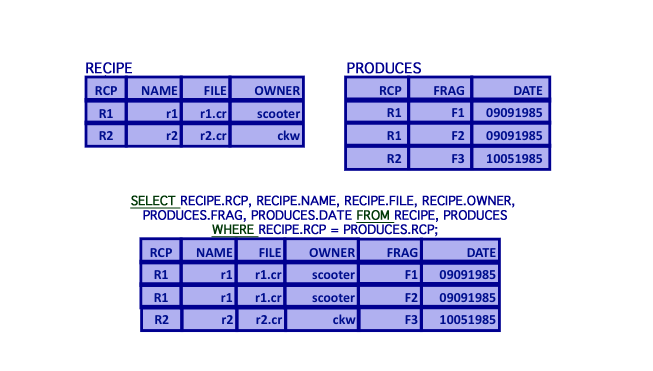
SELECT - Implicit Equijoin
- Join (equijoin)
- Matrix product of two relations based on equality of an attribute with the same domain
SQL - JOIN
- Joins two or more tables together based on a join predicate.
Note that the
JOIN keyword is actually part of theSELECT syntax - SYNOPSIS:
SELECT column1[,column2]FROM table1INNER JOIN table2ON join_predicate;SELECT column1[,column2]FROM table1INNER JOIN table2USING ( column_name) ;SELECT column1[,column2]FROM table1NATURAL JOIN table2;SELECT column1[,column2]FROM table1LEFT OUTER JOIN table2ON join_predicate; - Where:
- join_predicate is an equality for an Equijoin, or a comparison for any other join
SQL - INNER JOIN Examples
- Examples:
SELECT * FROM PRODUCES NATURAL JOIN;R1|F1|1985-09-09|r1|r1,cr|scooter R1|F2|1985-09-09|r1|r1,cr|scooter R2|F3|1985-10-05|r2|r2.cr|ckw
SELECT * FROM PRODUCES INNER JOIN RECIPE ON PRODUCES.RCP=RECIPE.RCP;R1|F1|1985-09-09|R1|r1|r1,cr|scooter R1|F2|1985-09-09|R1|r1|r1,cr|scooter R2|F3|1985-10-05|R2|r2|r2.cr|ckw
SELECT * FROM PRODUCES JOIN RECIPE USING(RCP);R1|F1|1985-09-09|r1|r1,cr|scooter R1|F2|1985-09-09|r1|r1,cr|scooter R2|F3|1985-10-05|r2|r2.cr|ckw
SQL - OUTER JOIN Examples
- Assume we add a new row (R3) into the RECIPE relation
- Outer join examples:
SELECT * FROM PRODUCES LEFT OUTER JOIN RECIPE ON PRODUCES.RCP=RECIPE.RCP;R1|F1|1985-09-09|R1|r1|r1,cr|scooter R1|F2|1985-09-09|R1|r1|r1,cr|scooter R2|F3|1985-10-05|R2|r2|r2.cr|ckw
SELECT * FROM RECIPE LEFT OUTER JOIN PRODUCES ON PRODUCES.RCP=RECIPE.RCP;R1|r1|r1,cr|scooter|R1|F1|1985-09-09 R1|r1|r1,cr|scooter|R1|F2|1985-09-09 R2|r2|r2.cr|ckw|R2|F3|1985-10-05 R3|r3|r3.cr|rst|||
SELECT -- Query Example
- Query: What recipes produce the AMP gene?
- First, select the AMP gene from the GENE relation and join it to CONTAINS
CREATE TABLE TEMP1 AS SELECT CONTAINS.FRAG,CONTAINS.GENE,GENE.NAME FROM CONTAINS,GENE WHERE GENE.ID=CONTAINS.GENE AND GENE.NAME="AMP"; - Note we're doing an implicit Equi-JOIN. To do the same thing more explicitly:
CREATE TABLE TEMP1 AS SELECT CONTAINS.FRAG,CONTAINS.GENE,GENE.NAME FROM CONTAINS INNER JOIN GENE ON GENE.ID = CONTAINS.GENE WHERE GENE.NAME="AMP";
- First, select the AMP gene from the GENE relation and join it to CONTAINS


SELECT -- Query Example 2
- Query: What recipes produce the AMP gene?
- Second, join the result to the PRODUCES relation and select the RCP attribute
CREATE TABLE TEMP2 AS SELECT PRODUCES.RCP FROM TEMP1,PRODUCES WHERE TEMP1.FRAG=PRODUCES.FRAG; - Note we're again doing an implicit Equi-JOIN. The explicit syntax would be:
CREATE TABLE TEMP2 AS SELECT PRODUCES.RCP FROM TEMP1 INNER JOIN PRODUCES ON TEMP1.FRAG = PRODUCES.FRAG;
- Second, join the result to the PRODUCES relation and select the RCP attribute


SELECT -- Query Example 3
- Query: What recipes produce the AMP gene?
- Finally, join the result to the RECIPES table
SELECT DISTINCT RECIPE.RCP, RECIPE.NAME, RECIPE.FILE, RECIPE.OWNER FROM TEMP2, RECIPE WHERE TEMP2.RCP = RECIPE.RCP;- Note the
DISTINCT keyword to remove duplicate rows
- Note the
- Finally, join the result to the RECIPES table


SQL - Query Example (shorthand)
- Most modern relational databases have good query optimizers
- Usually no need to create intermediate tables:
SELECT DISTINCT RECIPE.RCP, RECIPE.NAME, RECIPE.FILE, RECIPE.OWNER FROM GENE, CONTAINS, PRODUCES, RECIPE WHERE GENE.NAME = 'AMP' AND GENE.ID = CONTAINS.GENE AND CONTAINS.FRAG = PRODUCES.FRAG AND PRODUCES.RCP = RECIPE.RCP;
SQL - UPDATE
- Updates data in a database
- SYNOPSIS:
UPDATE tablenameSET columnname="newvalue"[,nextcolumn="newvalue2"...]WHERE columnnameOPERATOR "value" [AND |OR columnOPERATOR "value"]; - Example:
UPDATE GENE SET NAME='AMP' WHERE ID='G1';
- SYNOPSIS:
SQL - Other Useful Commands
ALTER - Alter a table after it has been created- Add or drop columns
- Add or drop primary or foreign keys
DELETE - Delete a row from a table. Syntax is similar toSELECT .DROP - Delete an entire table or database- SQL Functions - aggregation functions that operate on the results from a select
- Include basic statistics (
STDEV ,AVG ,SUM ,VAR ), and counting functions likeCOUNT (column) - Example:
SELECT COUNT(*) FROM RECIPE,PRODUCES WHERE RECIPE='scooter' AND RECIPE.RCP=PRODUCES.RCP;
- Include basic statistics (
SQL - References
- Good intro tutorial:
- On-line SQL Tutorial
- On-line SQL Tutorial 2
- Another good intro:
- A Gentle Introduction to SQL
- A useful reference site:
- W3 Schools SQL Tutorial
- Also worth a look
- Software carpentry lecture on Relational Databases
Object-Relational Databases
- Essentially an extension of the relational database model
- Preserves the tabular (relational) organization of the data
- Allows developers to define more complex data types (User Defined Types, UDTs)
- No support for encapsulation or inheritance
- Some support for methods is provided (User Defined Functions, UDFs)
- SQL object extensions already standardized (SQL3)
- e.g.
- postgres
- Oracle
Object Databases
- Provides persistent storage of objects
- Most useful in conjunction with object-based applications
- Primarily a programmer's tool, although vendors are providing SQL3 and ODBC interfaces
- e.g.
- Objectivity
Types of Databases
- Questions?
- Recommended Reading:
- Date, C.J. An Introduction to Database Systems. Reading, Mass.: Addison-Wesley (1981)
- Codd, E.F. A Relational Model of Data for Large Shared Data Banks. CACM 13, No. 6 (June 1970)
Uses of Databases
- ...or why do I [should you] care about this stuff?
- Three major computing issues in bioinformatics:
- Data processing -- convert raw data into results
- Visualization -- a way to visualize the results
- Data storage -- store the results (and perhaps the raw data)
Questions?
Database Access with Python
- SQL provides a way to interact with a relational database...
- ... but how do I access my database programmatically?
- Lots of ways, but we're going to discuss
sqlite3 - sqlite3:
- Provides access to
SQLite from python scripts - Simple (maybe too simple...)
- Basic idea is to
execute SQL commands and return the response as a python list - Installed on plato
- Provides access to
sqlite3 Example
#! /usr/bin/python
import sys
import sqlite3
try:
# Open a connection to the database
conn = sqlite3.connect ('bmi219.db')
cursor = conn.cursor()
# Execute an SQL statement -- can be pretty much any SQL
cursor.execute("SELECT NAME, PROTEIN from GENE")
# fetchall returns a list of lists
rows = cursor.fetchall()
for row in rows:
print "%s, %s"%(row[0], row[1])
# Close the cursor and commit any changes to the database
cursor.close()
conn.commit()
conn.close()
except sqlite3.Error, e:
# Handle any errors
print "Error %d: %s" % (e.args[0], e.args[1])
sys.exit (1)
AMP, MAKK... TET, MYAK... NGF, MYAK...
Larger sqlite3 Example
#! /usr/bin/python
import sys
import sqlite3
try:
conn = sqlite3.connect ('bmi219.db')
# Get a cursor we can work with
cursor = conn.cursor()
# Use the execute method to pass SQL commands to the database
cursor.execute("DROP TABLE IF EXISTS `GENE`")
# Note that we use triple quotes when we need multiple lines
cursor.execute("""
CREATE TABLE 'GENE'
(
'ID' char(16),
'NAME' varchar(20),
'PROTEIN' longtext,
'START' int,
PRIMARY KEY (`ID`)
)
""")
cursor.execute("""
INSERT INTO 'GENE' VALUES
('G1','AMP','MAKK...',-5)
""")
cursor.execute("""
INSERT INTO 'GENE' VALUES
('G2','TET','MYAK...',-10)
""")
cursor.execute("""
INSERT INTO 'GENE' VALUES
('G3','NGF','MYAK...',-1)
""")
print "Number of rows inserted: %d"%cursor.rowcount
# OK, now lets try to get some data out
cursor.execute("SELECT NAME, PROTEIN from GENE")
while (1):
row = cursor.fetchone ()
if row == None:
break
print "%s, %s"%(row[0],row[1])
print "Number of rows returned: %d"%cursor.rowcount
# Another way to do the same thing
cursor.execute("SELECT NAME, PROTEIN from GENE")
rows = cursor.fetchall ()
for row in rows:
print "%s, %s"%(row[0],row[1])
print "Number of rows returned: %d"%cursor.rowcount
cursor.close()
conn.commit ()
conn.close()
except sqlite3.Error, e:
print "Error %d: %s" % (e.args[0], e.args[1])
sys.exit (1)
SQLite3 Use
- sqlite3 provides a good low-level interface
- For most uses, probably want to wrap low-level SQL commands in Python objects
- In the above example, a
GENE might be an object - Might have methods to fetch (SELECT) or save (INSERT) a
GENE - Provides some insulation from underlying SQL implementation