Poster for NCRR Principle Investigator meeting June, 2004
Tom Goddard, Conrad Huang, Thomas Ferrin
Resource for Biocomputing, Visualization and Informatics
University of California, San Francisco
We are developing software for interactive visualization of virus capsids and other large molecular assemblies. Here we describe capabilities for visualizing large assemblies that are beyond what current molecular graphics programs provide. We also discuss data quality issues with virus structures that we are working with the Protein Data Bank to remedy. And we present approaches to making virus assemblies more user accessible via a variety of web technologies.
Current molecular graphics packages for analyzing individual macromolecules or small complexes perform well with up to a few hundred thousand atoms. Large virus capsids contain millions of atoms and run into memory and CPU speed limitations on desktop computers. Some optimizations allow responsive interactive visualization on such systems. In addition to optimizations, new capabilities such as cartoon representations for macromolecules and facilities for naming and navigating quaternary structure are basic ingredients for visualizing large assemblies.

|

|
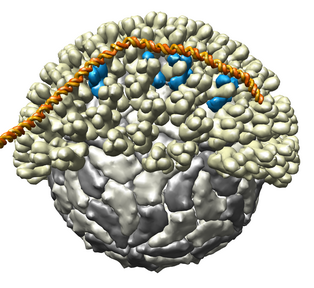
Figure 1. Bluetongue virus capsid with viral RNA stuck to outside of capsid. The capsid has 2 protein layers with half of the outer layer being cutaway in the picture. Atomic interactions between the capsid and RNA are shown. Residues involved in contacts of less than 5 angstroms are shown in black. These are PDB models 2btv and 1h1k.
Figure 1 shows an atomic resolution virus capsid structure containing 3 million atoms. Current molecular graphics programs use about 1 to 2 kilobytes of memory per atom. Building 3 gigabytes of data structures to open this model in a visualization program would take minutes. To avoid this high overhead we have developed code that only loads the atoms needed for atomic resolution display styles. Low resolution surfaces representing entire protein subunits can be shown using atomic information for just one copy of a multimeric subunit. This allows the structure shown to be loaded using modest amounts of memory (90,000 atoms for the unique subunits) in several seconds.
All contacts between the RNA (20,000 atoms) and capsid (3,000,000 atoms) were computed with a maximum range of 5 angstroms. A naive calculation of distances for all 60 billion atom pairs would take a few minutes on current desktop computers. A more sophisticated algorithm that places all atoms in 5 angstrom bins and limits comparisons to atoms in adjoining bins can be done in seconds.
These optimizations contribute to complexity in writing and maintaining the software and adding subsequent features. The calculation of atomic contacts has to account for the fact that not all atoms in the structure have been loaded into memory. It knows to use copies of atoms from structurally identical subunits to avoid loading atoms unnecessarily. These optimizations are relatively simple requiring a few thousand lines of code beyond the simple techniques used in molecular graphics programs aimed at small molecular systems.
|
|
|
|
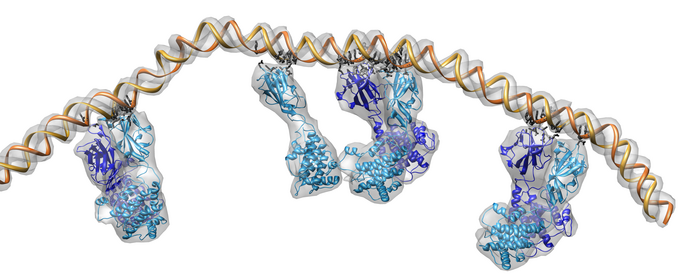
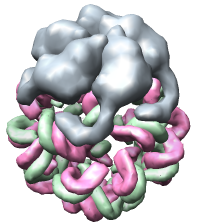
Figure 2. Satellite tobacco mosaic virus capsid with about 60% of viral single strand RNA genome. A single capsid protein makes contacts with many neighboring capsid proteins and several resolved segments of RNA.
Visualizing virus capsids requires capabilities not found in current molecular graphics software. The virus shown in figure 1 is studded with 780 copies of the outer capsid protein. Showing any substructure within these proteins provides an overwhelming level of detail when viewing the whole capsid. We have implemented a variable resolution surface depiction for sets of atoms that produces low resolution (e.g. 10 angstrom) cartoon depictions. Molecular surfaces used for studying individual macromolecules have much higher resolution and could not be used to render the virus in figure 1 at an interactive frame rate on even the fastest graphics hardware.
Most available virus capsid structures are icosahedral having an exact 60-fold symmetry. Sixty copies of the same atomic coordinates are used to form the full shell. The positions of the copies are specified using sixty 3 by 4 matrices each expressing a rotation and translation. Code to read these matrices and generate the multimeric structure is a basic requirement for visualizing large multimeric structures.
The virus capsid in figure 1 has two protein layers, the outer layer is made out of 13 slight structural variants of a single protein grouped in trimers, while the inner layer consists of two structural forms of another protein. Our software organizes the structure levels in a tree. The root of the tree is the full virus particle which has two children which are the inner and outer protein layers, the inner layer has children which are the 260 trimers and the leaves of the tree are the protein monomers.
The cutaway view in figure 1 was made by selecting the outer layer and then slicing it in half by dragging a box with the mouse. To select the outer layer a single protein is selected with the mouse and then promoted by pressing a button to select the containing trimer and again to select the outer layer. Without the hierarchical organization, selecting hundreds of individual proteins with a mouse would be extremely cumbersome. Another useful selection method is to promote a selection to all copies of currently selected subunits.
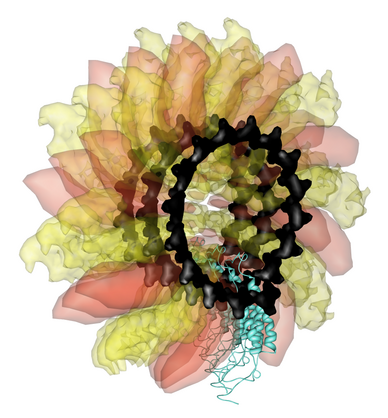
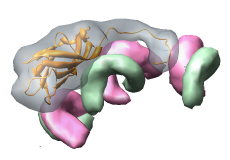
Figure 3. Ribgrass virus is helical. All available atomic resolution virus capsid structures are icosahedral or helical. |
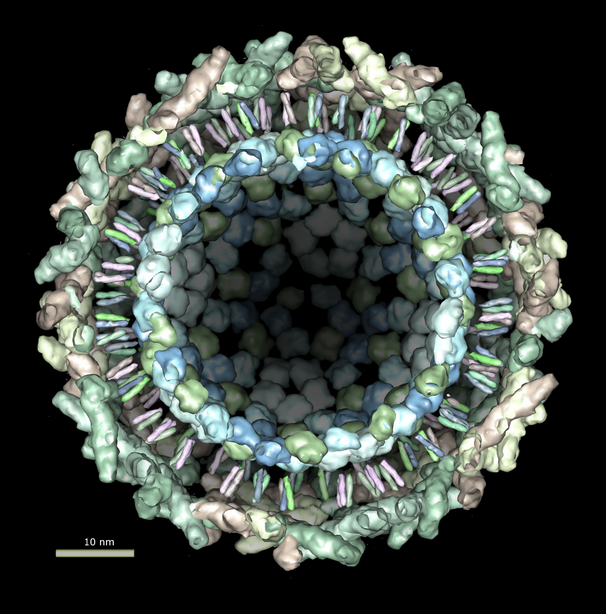
Figure 4. Sindbis virus has a lipid bilayer sandwiched between an inner protein nucleocapsid and outer glycoprotein layer. |
Our software for virus visualization has just basic capabilities, yet to our knowledge it is far more capable than any other distributed program. Because few programs can display virus capsids, the matrix data contained in the Protein Data Bank virus entries has not been extensively used. About half of the 200 virus entries do not describe the matrices needed to produce a full particle in a machine readable and correct form. We are working with the Protein Data Bank to identify the problem files and correct them.
It is difficult for researchers to explore the existing rich data set of over 200 virus structures (figures 2-5). Although our software, UCSF Chimera, has the basic virus display functionality, it requires reading extensive documentation to use it effectively. To make Chimera more accessible to researchers we are working with the Multiscale Modeling Tools for Structural Biology NCRR resource. They develop the Virus Particle Explorer (ViPER) web site which presents curated structure data for about 100 icosahedral viruses. We are trying three web technologies to let users explore this data beyond providing a fixed set of images: offering Virtual Reality Markup Language three dimensional models created by Chimera and viewable with a web browser plugin, launching our Chimera program directly from web browser links to show virus structures, and running Chimera on a web-server to render custom requested virus images in response to user settings specified in a browser.

Satellite Panicum Mosaic Virus, 1stm |

Satellite Tobacco Mosaic Virus, 1a34 |

Satellite Tobacco Necrosis Virus, 2stv |

Alfalfa Mosaic Virus, 1amv |

Cowpea Chlorotic Mottle Virus, ccmv_t2_672919 |

Densovirus, 1dnv |
Bacteriophage Fr, 1fr5 |
Bacteriophage FR, 1frs |
Bacteriophage MS2, 2ms2 |
Canine Parvovirus Strain D, 4dpv |
Bacteriophage ms2, 1aq3 |
Bacteriophage ms2, 1bms |
Feline Panleukopenia Virus Empty Capsid, 1c8e |
Bacteriophage GA, 1gav |
Porcine Parvovirus Capsid, 1k3v |
Bacteriophage Ms2, 1mst |
Feline Panleukopenia Virus, 1fpv |

Brome Mosaic Virus, 1js9 |

Murine Minute Virus, 1mvm |

Canine Parvovirus (CPV) Empty, 2cas |

Cowpea Chlorotic Mottle Virus, 1cwp |
Bacteriophage Qb, 1qbe |
Tobacco Necrosis Virus, 1tnv |
Adeno-Associated Virus, 1lp3 |

Tobacco Necrosis Virus, 1c8n |

Tobacco Ringspot Virus, 1a6c |
Bacteriophage PP7, 1dwn |

Red Clover Mottle Virus, 1rcmv |
Sesbania Mosaic Virus, 1smv |

Cucumber Mosaic Virus, 1f15 |

Rice Yellow Mottle Virus, 1f2n |
Cowpea Mosaic Virus, 1ny7 |
Turnip Yellow Mosaic Virus, 1auy |

Desmodium Yellow Mottle Virus, 1ddl |
Cocksfoot Mottle Virus, 1ng0 |
Southern Bean Mosaic Virus, 4sbv |
Human Papillomavirus 16 L1 capsid, 1dzl |

Tomato Aspermy Virus, 1laj |
Foot and Mouth Disease Virus (FMDV), 1bbt |
Bovine Enterovirus, 1bev |

Bean Pod Mottle Virus, 1bmv |
Physalis Mottle Virus, 1qjz |
Physalis mottle virus, 1e57 |
Human Rhinovirus 16, 1aym |
Echovirus 1 (FAROUK Strain), 1ev1 |

Human Rhinovirus 1A, 1r1a |

Human Rhinovirus 3, 1rhi |
Coxsackievirus B3, 1cov |
Echovirus 11 (Strain 207), 1h8t |

Chimeric Human Rhinovirus 14 w/ HIV1-V3 Loop, 1k5m |

Swine Vesicular Disease Virus, 1oop |

Human rhinovirus 14, 1r08 |

Human rhinovirus 14, 1rmu |

Human Rhinovirus 14, 4rhv |
Human Rhinovirus 2, 1fpn |

Theiler's Murine Encephalomyelitis (DA Strain), 1tme |

Mengo Encephalomyocarditis Virus, 2mev |
Cricket Paralysis Virus, 1b35 |
Poliovirus Type 1 (Mahoney Strain) at -170c, 1asj |
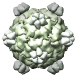
Bacteriophage G4, 1gff |

Poliovirus Type 1 (Mahoney Strain), 1hxs |
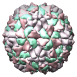
Poliovirus Type 3 (Sabin Strain), 1pvc |

Poliovirus Type 1 (Mahoney Strain) Empty Capsid, 1pov |
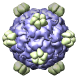
Bacteriophage phix174, 2bpa |

Human poliovirus 1 strain mahoney, 2plv |

Theiler's Murine Encephalomyelitis (BeAn Strain), 1tmf |
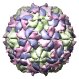
Poliovirus-2 Lansing Complexed with SCH48973, 1eah |

Cowpea Chlorotic Mottle Virus, Swollen Form (C), ccmv_swln_c |

Cowpea Chlorotic Mottle Virus, Swollen Form (D), ccmv_swln_d |
Black Beetle Virus, 2bbv |

d75n |
c74 |
Flock House Virus, 1fhv |

Flock House Virus, 2fhv |

Hepatitis B Virus, 1qgt |

Carnation Mottle Virus, 1opo |

Pariacoto Virus, 1f8v |
Bacteriophage phix174+scaffold, 1al0 |

Tomato Bushy Stunt Virus, 2tbv |

Nodamura Virus, 1nov |
Norwalk Virus Capsid, 1ihm |

Nudaurelia Capensis w Virus, 1nwv |

Sindbis virus, sindbis |
Human Rhinovirus 16 ICAM1 complex, 1d3e |
L-A Virus, 1m1c |
Simian Virus 40 (SV40), 1sva |
Murine Polyomavirus, 1sid |
Dengue virus, 1k4r |
pro1 |
Bacteriophage HK97 ProheadII, 1if0 |

Bacteriophage HK97 HeadII, 1fh6 |

Human Papillomavirus 16 L1, 1l0t |

Bacteriophage PRD1 Model, 1hb5 |

Bacteriophage PRD1, 1gw7 |

Bacteriophage PRD1 SUS607 mutant model, 1gw8 |

Bacteriophage PRD1 SUS1 mutant model, 1hb7 |

Bacteriophage PRD1 Wt Model, 1hb9 |

Bluetongue Virus, 2btv |
Bluetongue Virus inner layer, 2btv_t2 |
Rice Dwarf Virus, 1uf2 |
Rice Dwarf Virus inner layer, 1uf2_t2 |
Reovirus core, 1ej6 |
Reovirus core inner layer, 1ej6_inner |

Paramecium bursaria chlorella virus type 1, 1m4x28 |
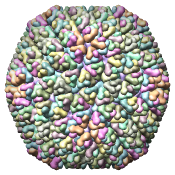
Figure 5. Automatically generated images of 100 virus capsids using data from the Virus Particle Explorer (ViPER) web site. Viruses are shown to scale. Different colors represent unique structural components (PDB chains).
The capabilities described in this poster have been implemented as the Multiscale extension to UCSF Chimera. Chimera is available on Windows, Mac, Linux, SGI and HP/Alpha computers. It is free for non-commercial use and can be downloaded at
http://www.cgl.ucsf.edu/chimera
The feature for calculating atomic contacts is not yet available as of our May 2004 release (Chimera 1.1951).
This work builds on the extensible UCSF Chimera molecular modeling system developed by Greg Couch, Tom Goddard, Dan Greenblatt, Conrad Huang, Elaine Meng, and Eric Pettersen. It is funded by the National Center for Research Resources grant P41 RR-01081.