The Selection Inspector
The Selection Inspector lists certain
attributes
of the currently selected items and allows
their values to be changed. The Selection Inspector can be opened:
- with Actions... Inspect in the menu
- by clicking the magnifying glass icon
 in the
status line
in the
status line
- by choosing Inspect from the context menu obtained by
double-picking
an atom, bond, or pseudobond (that is, doubleclicking with the button
assigned to picking)
Descriptions in the dialog are not necessarily identical to the
attribute names
(used in the code and in various
commands).
See also: attribute inspectors
Items available for inspection:
Write List... brings up a dialog for
saving a parsable text file of specifications
for the selected items.
Write PDB... brings up a dialog for
saving the selection as a PDB file.
Close dismisses the Selection Inspector, while
Help opens this manual page in a browser window.
The descriptions in the Selection Inspector
are not necessarily the same as the attribute names.
If the selection includes multiple items of the same type
and their attribute values vary, “-- multiple --” or
“ >1” will be reported for the attribute.
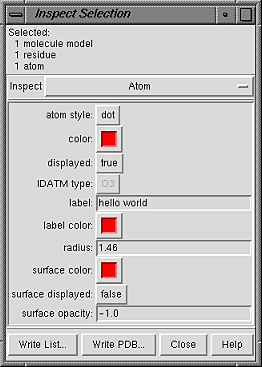 Atom attributes are:
Atom attributes are:
- atom style - atom draw mode
- color (a color well)
- color at the atom level;
see coloring hierarchy
- displayed - whether display is enabled at the atom level
(see display hierarchy)
- IDATM type - atom type
(currently shown in gray and cannot be changed)
- label - a text label for the atom.
If the atom label was shown using the menu or the
label command,
standard information (such as atom name) will be shown
in this field, but can be replaced arbitrarily;
however, showing the atom label again with the menu or
label
will restore the standard information.
- label color (a color well)
- if an atom label has no color of its own, it will inherit
the color of its associated atom; see
coloring hierarchy
- radius - radius in angstroms used when the
the draw mode is sphere; initially set
to an approximate VDW radius
- surface color (a color well)
- atom-level surface color; see
coloring hierarchy
- surface displayed - whether surface
display is enabled at the atom level
(see display hierarchy)
- surface opacity - atom-level surface opacity, ranging from
0 (completely transparent) to 1 (completely opaque); a negative value
indicates that the opacity included in the definition of the atom-level
surface color(s) should be used instead
Bond attributes are:
- bond style - bond draw mode
- color (a color well)
- color at the individual bond level (see
coloring hierarchy);
can also be controlled with the command
bondcolor.
Note that the color assigned to the bond will only be visible
when halfbond mode is off;
when halfbond mode is on, the color of each half of the bond
matches the visible color of the flanking atom.
- displayed - whether display is enabled at the individual
bond level (see display hierarchy);
can also be controlled with the command
bonddisplay
- true
- false
- if atoms shown
(displayed only when both flanking atoms are displayed)
- halfbond mode - whether the two halves of a bond are
handled as separate entities; when halfbond mode is on,
the color of each halfbond matches that of the attached atom
(can also be controlled with the command
bondcolor).
- label - an arbitrary text label for the bond
- label color (a color well)
- if a label has no color of its own, it will inherit
the color of its associated bond; see
coloring hierarchy
- radius - value to be multiplied by the molecule model
stick scale to generate
stick radius in angstroms (only applies to bonds in the stick
draw mode)
Residue attributes are:
- chi1 angle - protein/peptide sidechain
χ1 dihedral angle
- chi2 angle - protein/peptide sidechain
χ2 dihedral angle
- chi3 angle - protein/peptide sidechain
χ3 dihedral angle
- chi4 angle - protein/peptide sidechain
χ4 dihedral angle
- filled ring display - whether to fill rings
(applies only to rings of up to 6 members; see also
fillring)
- filled ring style
(see also fillring)
- thin
- thick - track the thickness of the surrounding bonds
- in helix
- in strand
- label - a text label for the residue.
If the residue label was shown using the menu or the
rlabel command,
standard information (such as residue name and number)
will be shown in this field, but can be replaced arbitrarily;
however, showing the residue label again with the menu or
rlabel
will restore the standard information.
- label color (a color well)
- if a residue label has no color of its own, it will inherit
the model color; see coloring hierarchy
- phi angle - protein/peptide backbone φ dihedral angle
(Ci-1-N-CA-C)
- psi angle - protein/peptide backbone ψ dihedral angle
(N-CA-C-Ni+1)
- ribbon color (a color well)
- residue-level ribbon color;
see coloring hierarchy
- ribbon cross section - ribbon
style
(see also ribrepr)
- ribbon display
- whether ribbon is shown;
ribbons are only drawn for proteins and nucleic acids.
Protein secondary structure assignments are taken from the input structure
file or generated with ksdssp.
- ribbon scaling - ribbon
scaling
(see also ribscale)
Molecule model attributes are:
- active - whether the model is
activated for motion
- aromatic color (a color well)
- color of aromaticity displays; No Color indicates matching
the corresponding atoms
- aromatic display
- whether to show aromaticity of displayed aromatic rings
(see also the command aromatic)
- aromatic line style
- line style of circles drawn to indicate aromaticity:
- aromatic ring style
- style of aromaticity display
(see also the command aromatic)
- auto-chaining
- whether to connect atoms that precede and follow undisplayed segments
(whether to draw pseudobonds between them)
- ball scale
- scale factor for atoms in the ball
draw mode.
The ball scale is multiplied by individual atom
VDW radii to generate ball radii in angstroms.
- color (a color well)
- color at the model level;
see coloring hierarchy
- displayed - whether display is enabled at the model level
(see display hierarchy)
- line style
- how to show bonds in the wire draw mode:
- line width - pixel width of lines depicting bonds
(when in the wire draw mode)
- residue label positioning
- which method is used to compute residue label position
(see rlabel for more details):
- centroid - geometric center of displayed atoms
- centroid of backbone - geometric center of displayed
backbone atoms, as defined by ribbon
residue
class (geometric center of displayed atoms for residues
without a residue class)
- primary atom - adjacent to the primary atom, generally
CA in an amino acid, C5' in a nucleotide, the first nonhydrogen atom in
residues of other types
- ribbon (cardinal) smoothing
- whether to perform additional smoothing of cardinal spline ribbon,
and if so, for which types of secondary structure
- none
- strand (peptide/protein strand)
- coil (peptide/protein nonhelix, nonstrand; nucleic acid)
- both (strand and coil)
- ribbon (cardinal) stiffness
- “resistance” to changes in the path direction of
cardinal spline ribbon (range 0.0-1.0)
- ribbon hides backbone atoms - whether showing ribbon for
residues within the model hides backbone atoms for those residues
(see the command ribbackbone
for details)
- ribbon inside color
(a color well)
- if assigned, this color will be used for the insides of all
protein helix ribbon segments in the model
regardless of how the rest of the ribbon is colored; No Color
restores the default behavior of matching interior and exterior
- ribbon spline
- method used to determine ribbon path (see also the command
ribspline)
- B-spline - path smoothed over five successive residues, may not
coincide exactly with any
mainchain
atoms
- cardinal - path smoothed over four successive residues, follows
guide
atom positions exactly unless additional smoothing
is applied
- silhouette
- per-model setting for silhouettes, outlines that emphasize borders and
discontinuities; the silhouettes will only be shown when the
global setting is also true
- stick scale
- scale factor for bonds in the stick
draw mode.
The stick scale is multiplied by individual bond radii (default
0.2 angstroms) to generate stick radii in angstroms.
Changing stick scale is preferable to changing all of the bond radii
in a model, because the former will also scale singleton atoms in the
endcap draw mode appropriately. Either way,
the other endcap atoms (those participating in bonds) will be scaled
to match the thickest of the attached bonds.
- surface color (a color well)
- model-level surface color; see
coloring hierarchy
- surface opacity - model-level surface opacity,
ranging from 0 (completely transparent) to 1 (completely opaque);
a negative value indicates that the opacity included in the definition
of the model-level surface color should be used instead
- vdw density - relative density of dots used in any VDW surfaces
(as with the command vdwdensity)
- vdw dot size - pixel size of dots in any VDW surfaces
Pseudobond
attributes are:
- bond style - pseudobond draw mode
- color (a color well)
- color at the individual pseudobond level; see
coloring hierarchy.
Note that the color assigned to the pseudobond will only be visible
when halfbond mode is off; when halfbond mode is on,
pseudobond color comes from the flanking atoms.
- displayed - whether display is enabled at the individual
pseudobond level
(see display hierarchy)
- true
- false
- if atoms shown
(displayed only when both flanking atoms are displayed)
- halfbond mode - whether the two halves of a pseudobond are
handled as separate entities; when halfbond mode is on,
the color of each halfbond matches that of the attached atom
(except if the bond style is spring,
one atom's color is used for the whole pseudobond)
- label - text label for the pseudobond;
if the pseudobond is a distance monitor, this label reports the distance.
Label text can also be assigned when a pseudobond is created with
PseudoBond
Reader.
- label color (a color well)
- if a label has no color of its own, it will inherit
the color of its associated pseudobond; see
coloring hierarchy
- radius - value to be multiplied by the
pseudobond group stick scale to generate
stick radius in angstroms (only applies to pseudobonds in the stick
draw mode)
Pseudobond group
attributes are:
- category
- name of the pseudobond group
- color
(a color well)
- color at the pseudobond group level; see
coloring hierarchy
- displayed - whether display is enabled at the pseudobond group level
(see display hierarchy)
- line style
- how to show pseudobonds in the
wire draw mode:
- line width - pixel width of
pseudobonds in the wire draw mode
- stick scale
- scale factor for pseudobonds in the stick
draw mode.
The stick scale is multiplied by individual pseudobond radii
(default 0.2 angstroms) to generate stick radii in angstroms.
MSMS surface attributes are:
- active - whether the model is
activated for motion
- color source - whether the surface color reflects
the color at the model or atom level
(see coloring hierarchy). When surface
color is set independent of these levels, unknown is reported.
This occurs when the surface is custom-colored, for example with
Surface
Color.
- displayed - whether surface
display is enabled at the model level
(see display hierarchy)
- dot size - pixel size of dots used in the dot
surface representation
- line width - pixel width of lines used in the mesh
surface representation
- pieces
- number of surface pieces
(independently selectable subparts) in the model
- probe radius
- radius in Å of the probe sphere used to compute the surface.
A larger probe decreases surface bumpiness because it fits into fewer crevices.
A radius of 1.4 Å is commonly used to approximate a water molecule.
- representation - which type of
surface representation
is being used
- selectable with mouse
- whether the surface piece(s)
comprising the model can be
picked with the mouse
- show disjoint surfaces - whether to check
for multiple disjoint surfaces rather than assuming there is only one
(increases calculation time)
- silhouette
- per-model setting for silhouettes, outlines that emphasize borders and
discontinuities; the silhouettes will only be shown when the
global setting is also true
- triangles - number of triangles in the surface model
- vertex density - vertices per Å2
used to compute the surface.
Greater density results in a smoother surface but
increases computational demands for calculating and moving the surface.
Surface piece attributes are:
- color (a color well)
- surface piece color
- display style - type of surface display
- filled
(sometimes simply called
surface)
- mesh
- dot
- displayed - whether the surface piece is displayed
(see display hierarchy)
- dot size - pixel size of dots in the dot display style
- lighting - similar to
mesh
lighting except affecting both mesh and filled styles
- lighting, transparency - whether to
dim
transparent surfaces
- lighting, two sided - whether
two-sided lighting is in effect
- line smoothing - whether to
smooth lines in the mesh display style
- line thickness - pixel width of lines in the mesh
display style
- triangles - number of triangles in the surface piece
Surface (SurfaceModel) attributes are:
- active - whether the model is
activated for motion
- displayed - whether display is enabled at the model level
(see display hierarchy)
- pieces
- number of surface pieces
(independently selectable subparts) in the model
- selectable with mouse
- whether the surface piece(s)
comprising the model can be
picked with the mouse
- silhouette
- per-model setting for silhouettes, outlines that emphasize borders and
discontinuities; the silhouettes will only be shown when the
global setting is also true
- triangles - number of triangles in the surface model
UCSF Computer Graphics Laboratory / November 2013
 Atom attributes are:
Atom attributes are: