August 20, 2007
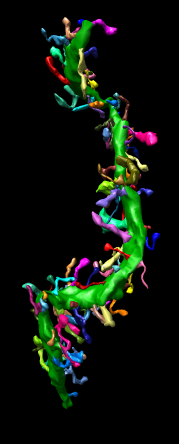
This extension allows Chimera to read IMOD binary files containing segmentation meshes and display them as a surface in Chimera. The mesh for each object in the IMOD file becomes a separate surface group in a single Chimera surface model and is colored as indicated in the IMOD file. This file reader only displays IMOD meshes. It can be reconfigured to also show contours (layerlines) and sets of points.
Several keyboard accelerators allow changing surface display. To enable keyboard accelerators use menu entry Tools / General Control / Accelerators On.
These commands act on selected objects. To select an object click on it with the left mouse button while holding the ctrl key down. Multiple objects can be selected by dragging a box around them using ctrl-left-mouse-button. Holding down the shift key while selecting will add new items to the selection, or unselect already selected items. Ctrl-clicking on the background (i.e. on no object) clears the selection.
Mesh Display. Selected objects can be displayed as triangular meshes using keyboard accelerator "ms" (mesh surface). Accelerator "fs" (filled surface) returns selected objects to surface display style.
Coloring. Selected objects can be given new colors using accelerator "co" which will display a color chooser dialog.
Area and Volume. The surface area and enclosed volume of selected segmented object can be measured using accelerators "ma" (measure area) and "mv" (measure volume) (available in Chimera version 1.2297 and newer). If the surface for an object contains holes, then no volume is reported.
The name of a segmented object is shown in a balloon when the mouse is hovered over that object. This is broken in Chimera 1.2297 and earlier, the balloon shows nothing unless a molecule is open in Chimera.
Contour lines and sets of points can be displayed. By default, only mesh data in the IMOD file is read and displayed using a surface model. Creating contours is slow so they are not read unless a modification is made to the code to enable them. Contour lines and points are displayed using Chimera volume path tracer markers and links (implemented as molecule atoms and bonds). For each IMOD object containing contours a separate marker set is created. Each marker set is a separate model in Chimera.
To enable contour reading edit the file
changing "contours = False" to "contours = True" and restart Chimera.
No session saving. IMOD surfaces are not saved in Chimera session files. You can change the colors of IMOD surface objects using keyboard shortcut "co" but there is no way to save the new colors in Chimera.
August 20, 2007. Documented lack of of session saving.
May 11, 2007. Placed contours for each object in a separate marker set. Formerly all contours and points were placed in a single marker set and which objects they belonged to was lost.
May 10, 2007. Added support for reading contours and points from the IMOD file, creating a volume path tracer madel.
February 27, 2007. Updated for Chimera version 1.2348 which uses NumPy instead of Numeric Python.
September 29, 2006. Created file reader for Masako Terado and Maryann Martone of the National Center for Microscopy and Imaging Research (NCMIR) to assist in measuring lengths of dendritic spines within Chimera.