
 |
|
Transparency1
 JPEG
version (105KB), TIFF version
(1,168KB)
JPEG
version (105KB), TIFF version
(1,168KB)
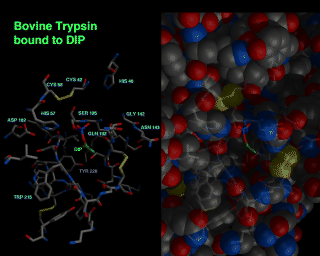
This shows the active site of trypsin, with the inhibitor DIP bound to the active site. In order to get a feel for the active site pocket, as well as the residues involved in binding, a side by side view, with faint residues in stick fashion combined with space filling conic on the right and just the stick representation on the left was used.
- MidasPlus was opened as a half screen, as the final picture will be a side by side image of two half screens. This happens automatically when MidasPlus is opened by starting the window as high in the left hand upper corner as it will go on the screen, and then dragging the mouse to the bottom left corner of the screen.
- The appropriate view of the active site of the molecule was chosen. Save the position, in case you accidentally move the protein later: command: savepos mypicture
- The spheres displayed on the screen are saved into a file, but the inhibitor is undisplayed, as it will not be displayed as spheres: command: ~display :inh (inh = name of inhibitor residue) command: pdbrun cat > spheres.pdb
- The inhibitor is put into a file containing the "sticks" representation available using neon. Everything on the screen is undisplayed, except the inhibitor. Then "stick" coordinates are written into a file. command: show :inh command: preneon cat > sticks.pdb
- A conic image of this file is made. Outside of MidasPlus, the conic program is run on a file composed of the two above files. Combine these files and run conic: cat spheres.pdb sticks.pdb > all.pdb conic -o part1.pic all.pdb
- If you are happy with part1.pic, you can get rid of the large spheres.pdb and sticks.pdb files. You can look at it with ilabel or ipaste: ipaste part1.pic Now make the right side of the image containing the key residues and inhibitor as sticks. Go back to your MidasPlus window. YOU HAVE BEEN CAREFUL NOT TO MOVE YOUR PROTEIN AROUND, as everything must be aligned perfectly. If you have moved things, reset to your original position with the reset command: command: reset mypicture Now show all the key residues of interest only. If there are a lot of them, you might have them listed in a file, such as: display :14, 15, 117, 243, inh By sourceing a file containing this command in it, you can have your desired key residues on the screen: ~display source displayfile
- Make a neon image of this file. See the neon man page for details on controlling the types of sticks, thickness, etc., using the "neon.dat" file. command: neon -o leftside.pic
- In order to get the "blended" molecular skelton imposed on the spheres, now use the SGI utility "blend". At this point, you are done with MidasPlus. blend leftside.pic part1.pic blended.pic .5 This creates a new picture, blended.pic, with ghostly bonds inside, and a stong inhibitor showing on the outside, plus the spheres representing the protein.
- To assembe the two sides of the slide, leftside.pic and blended.pic, together, the SGI utility "assemble" is used. assemble 2 1 final.pic leftside.pic blended.pic See the man page on "assemble" for more details. View the final image with ipaste, and no borders: ipaste -n final.pic
DIP trypsin structure solved by J.L. Chambers, R.M. Stroud, and J. Finer-Moore. Image by Julie Newdoll. ©2004 The Regents, University of California; all rights reserved.