This tutorial focuses on looking at sequences and structures together using the Multalign Viewer extension. Members of the enolase superfamily of enzymes are used to illustrate these features of Chimera. The enolase superfamily has been described in several publications, including:
P.C. Babbitt, M.S. Hasson, J.E. Wedekind, D.R. Palmer, W.C. Barrett, G.H. Reed, I. Rayment, D. Ringe, G.L. Kenyon, and J.A. Gerlt, "The Enolase Superfamily: A General Strategy for Enzyme-Catalyzed Abstraction of the Alpha-Protons of Carboxylic Acids" Biochemistry 35:16489 (1996).
To follow along with the tutorial, you will first need to download the MSF file super8.msf into your working directory. This file contains a sequence alignment of the barrel domains of eight enolase superfamily members.
On Windows/Mac, click the chimera icon; on UNIX, start Chimera from the system prompt:
unix: chimera
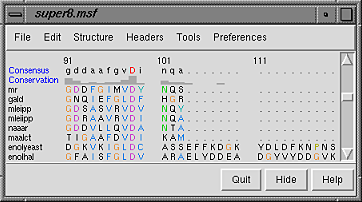
A basic Chimera window should appear after a few seconds. Choose the menu item Tools... Structure Comparison... Multalign Viewer. In the resulting dialog, locate and open super8.msf (the file type is MSF). A Multalign Viewer window containing the alignment will appear. Multalign Viewer has its own preferences; choose Preferences... Layout from the Multalign Viewer menu and change the font size and the sequence-wrapping behavior as desired. Size and place the sequence window and main Chimera window so that both are visible. If the sequence window becomes obscured at any point, it can be raised from the Chimera Tools menu (Tools... MAV - super8.msf... Raise).
| Multalign Viewer sequence window |
|---|
 |
When the mouse focus is in the sequence window, the Page Down key (or space) moves the alignment view down and Page Up (or Shift-space) moves the alignment view up.
The names of the sequences are shown on the left; a Consensus sequence and Conservation histogram are shown above the multiple alignment. The superfamily is very diverse; there are very few positions in the alignment in which all the sequences have the same residue (indicated with a red capital letter in the consensus sequence).
The first sequence in the alignment, named mr, corresponds to the barrel domain of mandelate racemase from Pseudomonas putida. The next-to-the last sequence in the alignment, named enolyeast, corresponds to the barrel domain of enolase from Saccharomyces cerevisiae. There are multiple structures in the Protein Data Bank for each of these sequences; 2mnr (mandelate racemase) and 4enl (enolase) are used in this tutorial.
To open the structures, choose File... Fetch by ID from the Chimera menu. In the resulting dialog, check the PDB ID option (if it is not already checked) and the option to Keep dialog up after Fetch. Enter 2mnr in the blank marked PDB ID and click Fetch. Repeat the process to fetch 4enl, and then click Close to dismiss the dialog. Chimera will attempt to find the files within a local installation of the Protein Data Bank. If a file is not found locally, Chimera will try to retrieve it from the Protein Data Bank web site. If this procedure does not work, it may be that you do not have internet connectivity; instead, download the files 2mnr.pdb and 4enl.pdb included with this tutorial and open them in that order as local files (with File... Open).
The view is initially centered on a protein colored white, the mandelate racemase structure. Off to the side (and possibly out of view) is the enolase structure, which is colored magenta. Readjust the view to focus on both structures:
Actions... FocusThe structures are not matched in any way; the coordinates are taken straight from the Protein Data Bank. Rotate, translate, and scale the structures as needed to get a better look (see mouse manipulation to review how this is done). If you like, open the Side View; choosing Tools...Viewing Controls... Side View is one way to do this. Continue moving and scaling the structures as desired throughout the tutorial.
| sequence window after structure association |
|---|
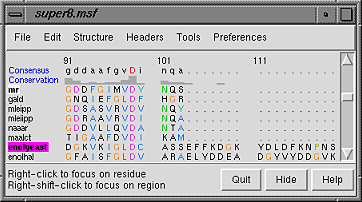 |
Notice that in the sequence window, the sequence names mr and enolyeast are now shown in bold within boxes colored white and magenta, respectively. This means that the sequences have been compared with the sequences in the structures, found to match sufficiently well, and automatically associated with the structures. The colors indicate which sequence has been associated with which structure.
The alignment includes just the barrel domains, so it will be simplest to display only a chain trace of these portions. First, open the Command Line; choosing Tools... General Controls... Command Line is one way to do this. Next, find out which residue numbers in the structures correspond to the beginning and end of the alignment. In general, these numbers will not be the same as the numbers marked on the alignment. However, placing the cursor over any residue in a sequence that is associated with a structure gives the corresponding structure residue number near the bottom of the sequence window. In each of the two structure-associated sequences, find out the starting and ending residue numbers by placing the cursor over the first and last residues in the alignment. Doing this reveals that the residue ranges are 134-317 in the mandelate racemase structure and 151-400 in the enolase structure. Display the alpha-carbon chain traces of the barrel domains and increase the linewidth (review atom specification syntax if desired):
Command: chain #0:134-317@ca
Command: chain #1:151-400@ca
Command: linewidth 2
The sequence alignment can be used to guide a structural match. From the Multalign Viewer menu, choose Structure... Match... and make 2mnr the reference structure and 4enl the structure to match. Click Apply without checking any boxes. This causes the match to use all pairs of alpha-carbons of residues aligned in the sequence alignment. Readjust the view to focus on the structures:
Actions... FocusThe match is fairly rough; the RMSD is 8.4 angstroms (match values are written to the Reply Log, Tools... Utilities... Reply Log). It is evident that not all of the residues aligned in the sequence alignment are really structurally equivalent. Some loops in enolase (magenta) are much longer than those in mandelate racemase (white). Try the structure matching again, but this time, check the box marked Iterate by pruning... and edit the angstrom value to 1.0 before clicking OK. This will superimpose only the pairs that collectively match very well in space. Visually, the match is improved; the dissimilar loops were not used in the match.
Next, we will see where some of the conserved residues are within the structures. In the sequence window, use the mouse to drag a column containing the first completely conserved residue in the alignment (the aspartate, D, at alignment position 99). This selects the residues in the associated structures, which then can be displayed:
Actions... Atoms/Bonds... showOpen the Region Browser (Tools... Region Browser in the Multalign Viewer menu). If a sequence region is created by mistake, it can be deleted by clicking on its line and then Delete within the Region Browser.
| active sites |
|---|
 |
Create another region (this time, press Ctrl along with the mouse button to start a new region) for the next completely conserved residue. This is a glutamate, E, at alignment position 148; display it too:
Actions... Atoms/Bonds... showThe displayed residues point into the center of the barrel, ligating a catalytically important metal ion. Display the active site metal ions:
Select... Structure... ionsSequence regions can be created automatically. From the Multalign Viewer menu, choose Structure... Secondary Structure... show actual. This creates regions in the structure-associated sequences named structure helices (light yellow with gold outline) and structure strands (light green with green outline). The region names are listed in the Region Browser. Clicking on a region in the sequence window (or clicking the Active checkbox for that region in the Region Browser) will select the corresponding residues in any associated structures. For sequences not associated with structures, Structure... Secondary Structure... show predicted creates regions named predicted helices and predicted strands shown with gold and green outlines, respectively. The GOR method is used for prediction. Close the Region Browser if it is open.
Actions... Atoms/Bonds... show
Select... Clear Selection
| conservation shown with color |
|---|
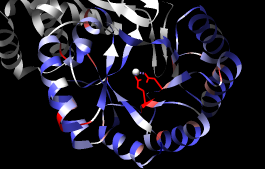 |
A structure can be colored according to the conservation in an associated sequence alignment. First, close one of the structures, clear any selections, and show a ribbon for the other structure:
Command: close 1Choose Structure... Render by Conservation from the Multalign Viewer menu. The resulting Render by Attribute tool shows a histogram of the residue attribute mavPercentConserved, the percent conservation of the most prevalent residue at the corresponding position in the alignment. Adjust the coloring sliders on the histogram (and their Color values, if desired) before clicking Apply. Coloring the structure by mavPercentConserved shows the high conservation of the metal-binding residues and the low conservation of most residues around the outside of the barrel.
Command: ~select
Command: ribbon
The residue attribute mavConservation is also listed in the Render by Attribute dialog. Its values are those shown in the Conservation line in the sequence window. Several different methods for calculating conservation are available. The Multalign Viewer Analysis preferences (Preferences... Analysis) control which method is used. If you wish, change the Conservation style to AL2CO and see how the Conservation histogram in the sequence window changes as parameters are varied. It is necessary to use Refresh... Values in the Render by Attribute tool to update the values in the histogram before recoloring a structure with Apply or OK (the latter will also close the tool). When finished, Close the Multalign Viewer preferences and Render by Attribute.
See the Multalign Viewer documentation for a full description of its many functions, including alignment editing. A number of sequence alignment formats can be read in and (with or without prior editing) written out.
When finished, end the Chimera session:
Command: stop