
Boltz single sample, Mac vs Linux

Boltz 1 sample vs 2 samples
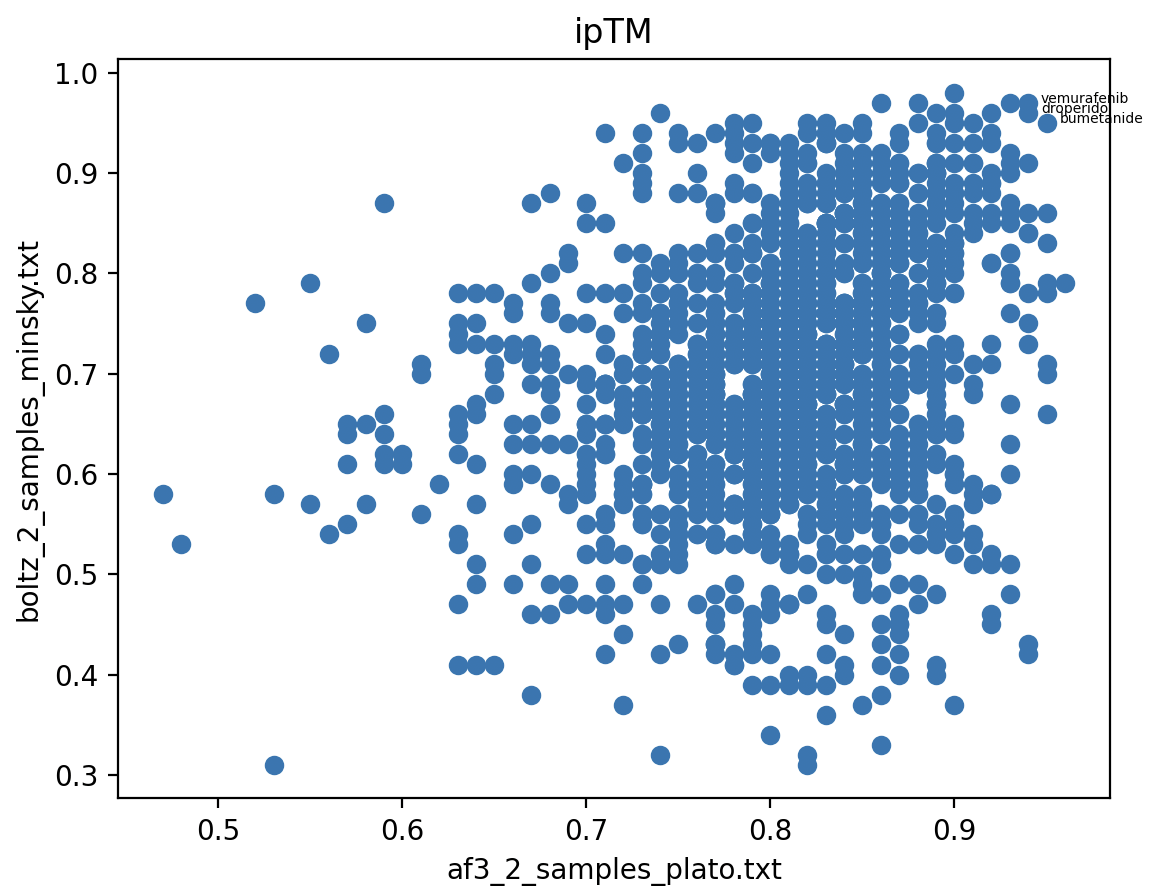
AlphaFold 3 vs Boltz
Tom Goddard
February 13, 2025
We compare predictions of 1508 FDA drug active ingredients bound to monkeypox E10R protein (mRNA decapping enzyme) made with Boltz-1 and AlphaFold 3. I ran 3 sets of Boltz-1 predictions: one on Mac Studio M2 Ultra (descartes, 25 hours), one on Linux Nvidia 4090 (minsky, 3.5 hours), both of those predicting just 1 structure for each drug, and one set on Linux Nvidia 4090 predicting 2 structures per drug (minsky, 4.5 hours). I ran one set of AlphaFold 3 predictions predicting 2 structures per drug (wynton, 7 hours). Another page explains how the AlphaFold3 runs were done and details for making the runs on the UCSF Wynton cluster. The Boltz predictions on Mac M2 Ultra took about 7x longer than Linux Nvidia 4090 (about 1 minute per drug vs 8 seconds), and the GPU power dissipation on the Mac was 40 Watts (measured with powermetrics command), while the Nvidia power was 400 Watts (measured with nvidia-smi command).
Scatter plots of ipTM (interface predicted TM scores) show Boltz-1 runs gives similar ipTM scores for each drug across the 3 runs. The scatter plot of AlphaFold 3 ipTM score versus Boltz-1 ipTM score (best of 2 predicted structures) show very different scores from the two methods with a rather weak correlation (ie high scoring drug with Boltz is not often high scoring with AF3).

Boltz single sample, Mac vs Linux | 
Boltz 1 sample vs 2 samples | 
AlphaFold 3 vs Boltz |
Here are 3 drugs that have very high ipTM scores (>= 0.94) in both Boltz and AlphaFold 3 predictions. We see the drug pose is very similar in the first two and roughly similar in the third. It is a bit challenging to give RMSD values for the drug atoms because Boltz and AlphaFold 3 name all the atoms differently.
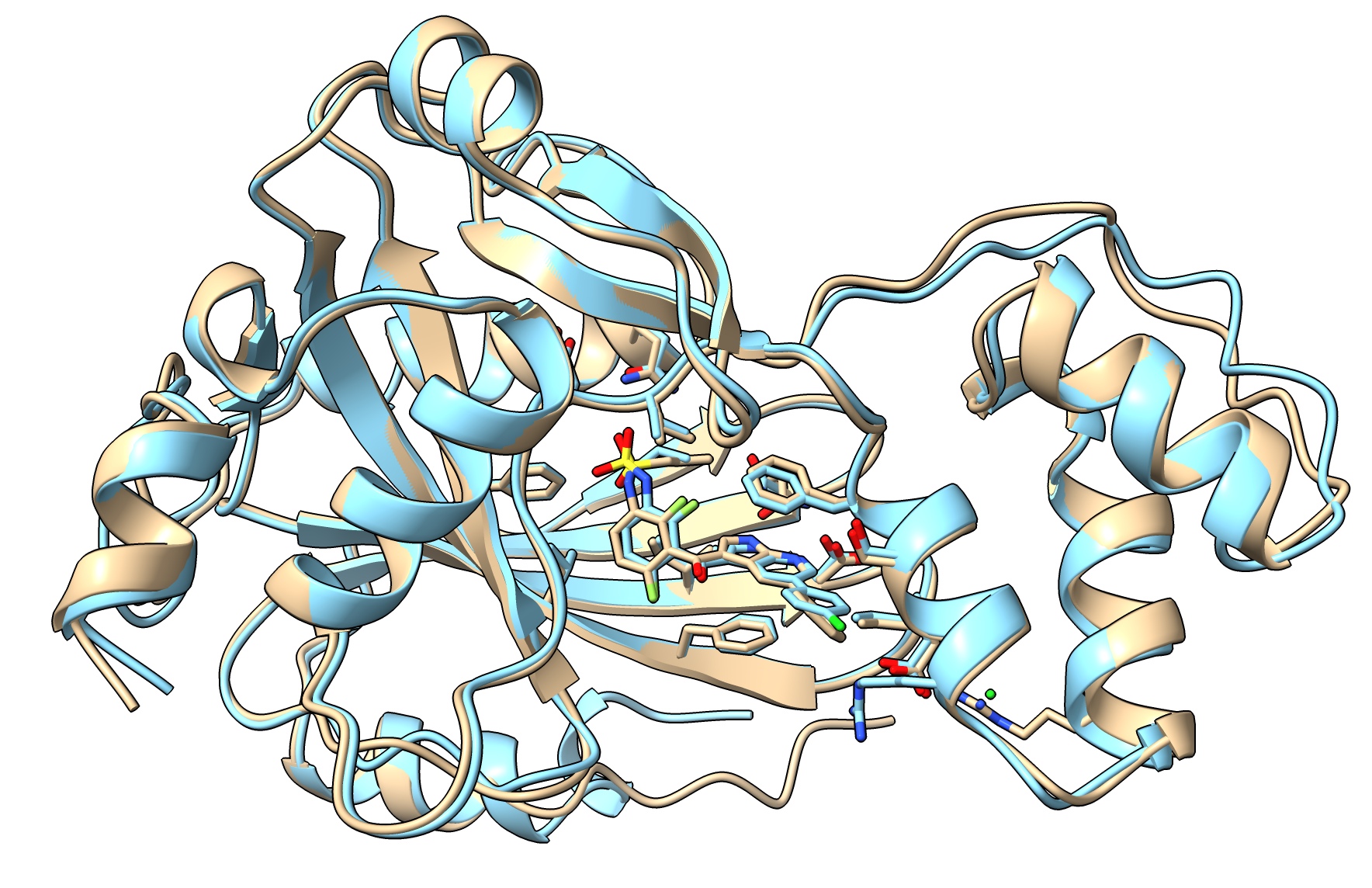
Vermurafenib AlphaFold 3 (tan), Boltz-1 (blue) | 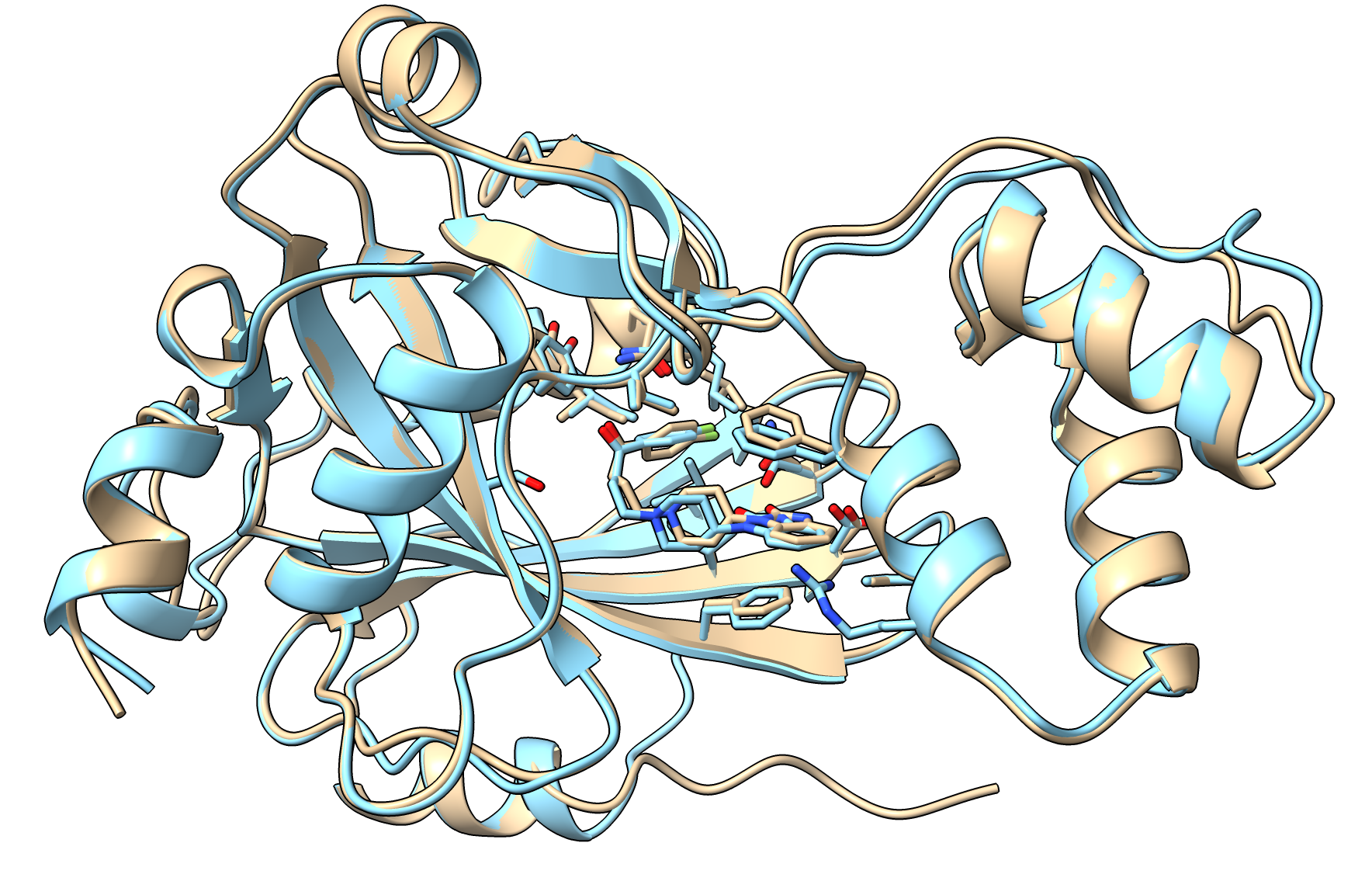
Droperidol AlphaFold 3 (tan), Boltz-1 (blue) | 
Bumetanide AlphaFold 3 (tan), Boltz-1 (blue) |
ChimeraX 1.9 session files for these 3 high scoring predictions are here vemurafenib_af3_boltz.cxs and droperidol_af3_boltz.cxs and bumetanide_af3_boltz.cxs.
Here are ipTM, pLDDT, and PAE scores for the 4 sets of predictions.
I have not included links to the predictions on this web page since since it is about 6 Gbytes of .cif and PAE files, but I can give you those on request. The scores were calculated from the prediction .cif and PAE files using ChimeraX scripts af3_drug_scores.py and boltz_drug_scores.py invoked using ChimeraX without its graphical interface like this
chimerax --nogui --exit --cmd "run af3_drug_scores.py */*_model.cif" >& drug_scores_af3.txt &
I wrote this Python script scatter.py with matplotlib to make the scatter plots.