ChimeraX graphical user interfaces for Phenix tools
Tom Goddard
January 30, 2025
Phenix R24 advisory meeting
R24 Aim 2:
Improve program usability and integration with other community software resources
...including an interface to UCSF's ChimeraX, a visualization and analysis program widely
used by the cryo-EM community...
UCSF Team Members: Tom Ferrin, Tom Goddard, Elaine Meng and Eric Pettersen
ChimeraX
- Used by biology researchers worldwide, including almost all cryoEM labs. ChimeraX web site.
- Free for academics, paid commercial license for pharmaceutical companies.
- For interactive visualization, analysis and building of macromolecular complexes (proteins, nucleic acids, ligands).
ChimeraX Use Statistics
- ChimeraX is replacing our previous Chimera visualization software, about half of users have updated.
- 2571 journal citations in 2024, graph.
- 6500 new voluntary registrations in 2024.
- 15379 new registrations in last 3 years, database (password required).
- 5537 unique IP addresses downloaded the ISOLDE model refinement ChimeraX extension in 2024.
- 950 unique IP addresses downloaded the PhenixUI ChimeraX extension in 2024.
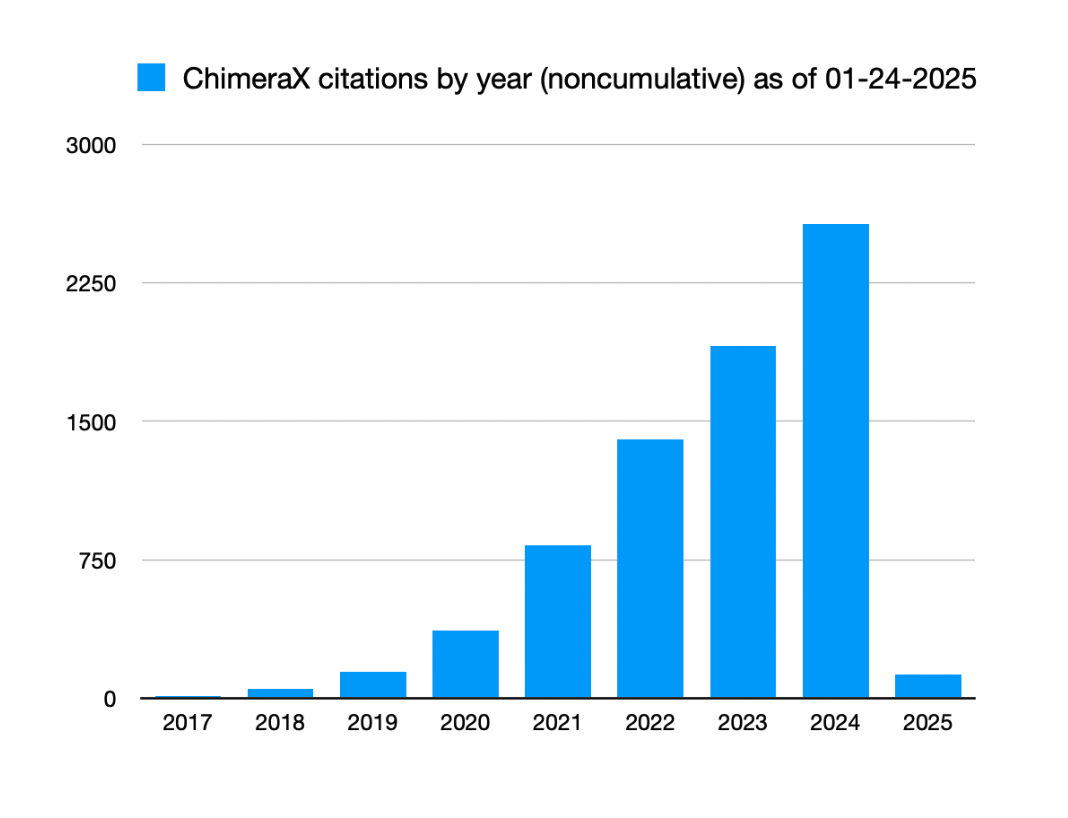
Journal citations per year
| 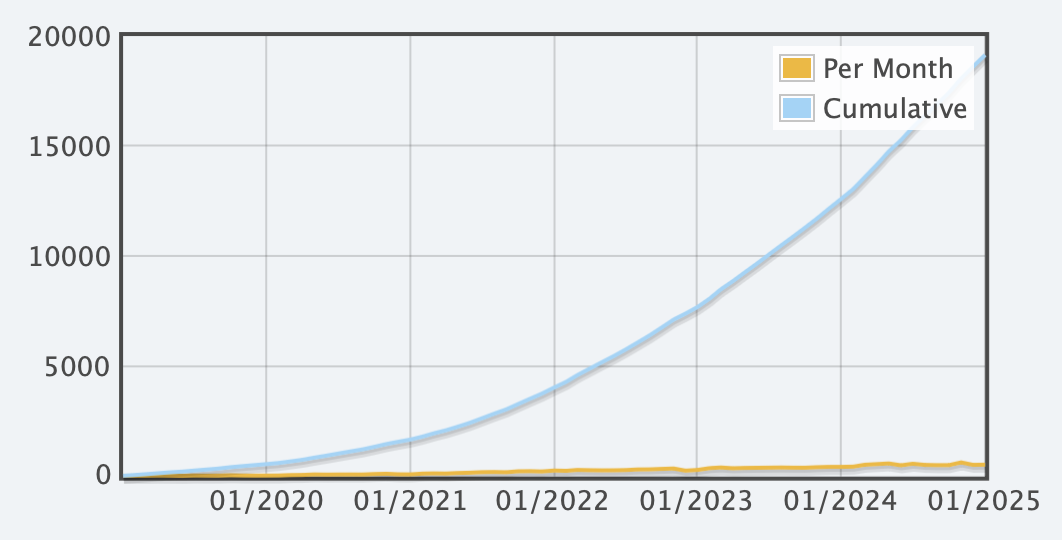
Registrations, voluntary, cumulative
| 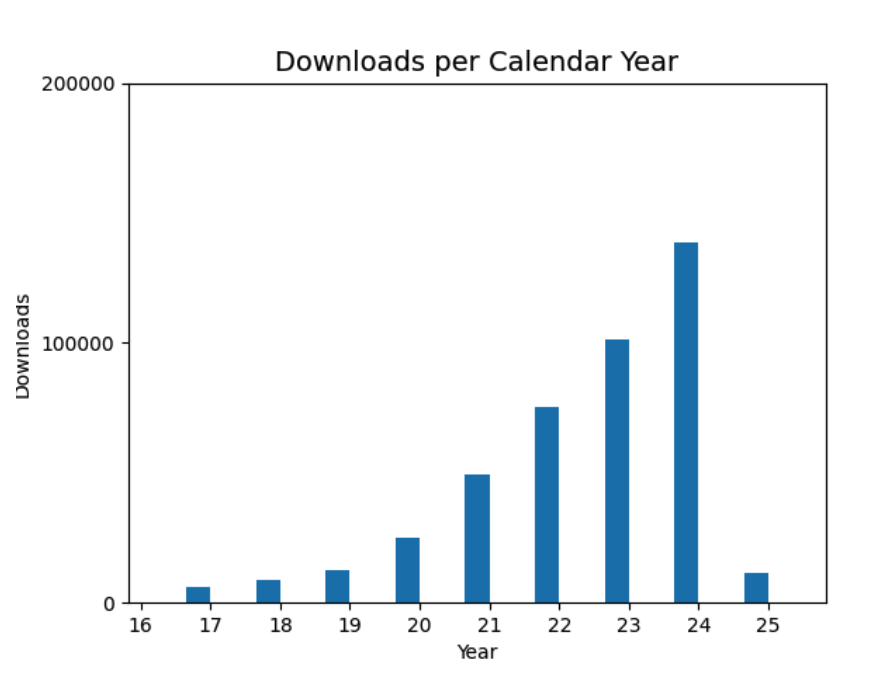
Downloads per year
|
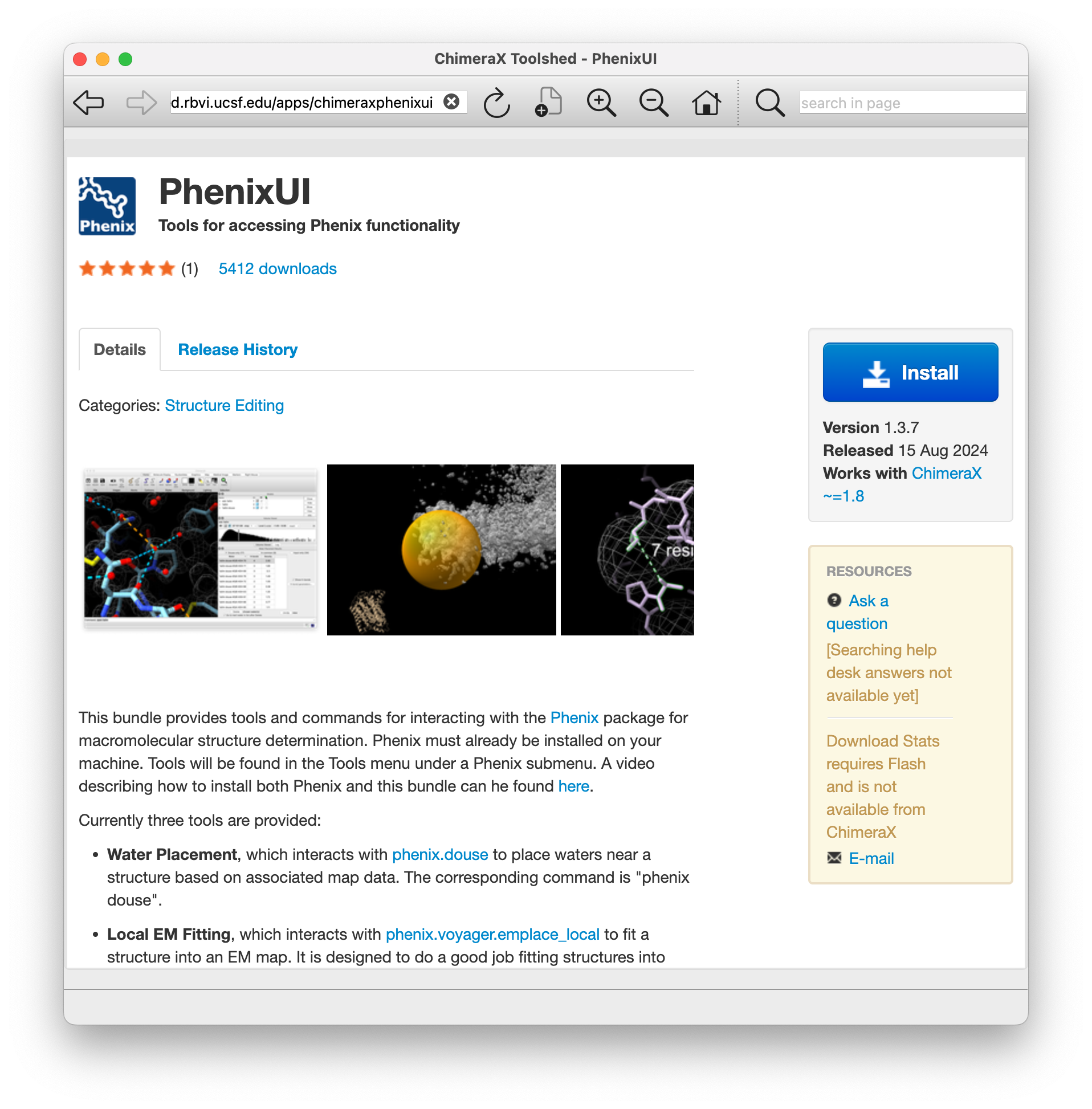
Obtaining ChimeraX User Interfaces for Phenix Tools
|
- ChimeraX extensions are distributed through the ChimeraX Toolshed web site.
- Users are notified if updates are available when ChimeraX starts.
- PhenixUI Toolshed web page shown at left.
- Both ChimeraX and Phenix must be installed on the researcher's computer.
- Tutorial video How to use Phenix with ChimeraX by Dorothee Liebschner (2 min 28 seconds).
|
Current ChimeraX Phenix User Interfaces
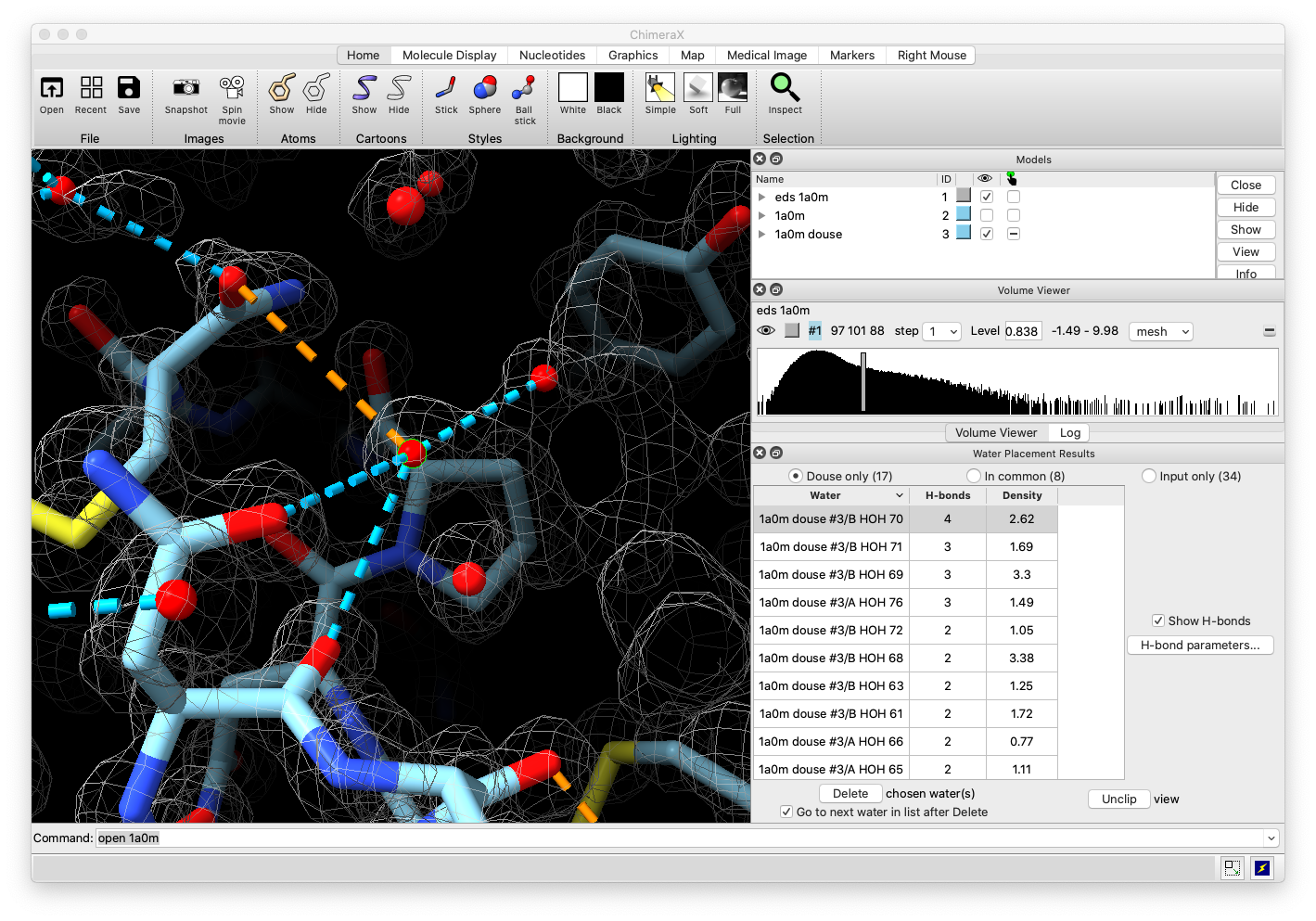
Place waters
| 
Local EM fitting
| 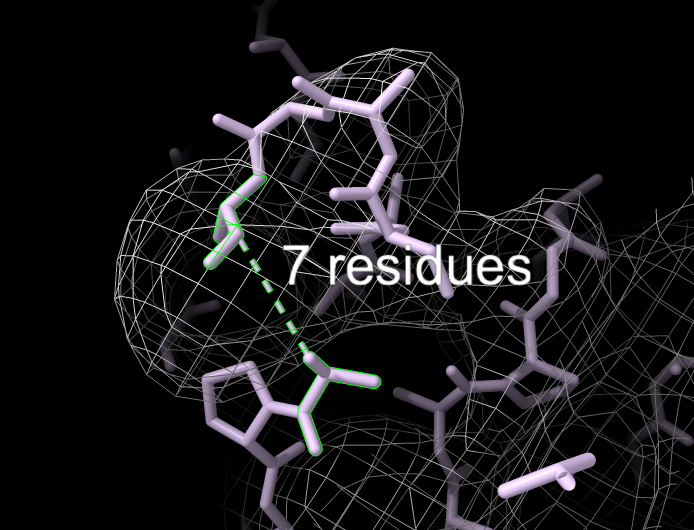
Fit loops
|
- Water placement in cryoEM density maps.
- Local EM fitting of atomic models in cryoEM maps.
- Fit loops - adds loops of up to 15 residues to an existing atomic model following density.
- Fit ligand - place known ligand in chosen map density (development in progress).
ChimeraX User Interface Development Process
- ChimeraX Phenix user interfaces are developed at UCSF by Eric Pettersen.
Extensive documentation is written Elaine Meng. Tom Goddard advises on cryoEM issues.
- Phenix developers Randy Read, Tom Terwilliger, Pavel Afonine, Oleg Sobolev, Billy Poon
make Phenix improvements to address any issues uncovered.
- Dorothee Liebshner creates tutorial videos.
- Monthly meetings including the Phenix and ChimeraX teams.
- Demonstrate current tool interfaces.
- Take suggestions for user interface improvements.
- Discuss modifications to Phenix command to support better user interfaces.
- Discuss Phenix command issues discovered in testing.
- Choose next Phenix tool to interface with ChimeraX.
User Interfaces
- Atomic models and cryoEM or X-ray maps are opened in ChimeraX.
- Phenix tools are chosen from the Tools/Phenix menu.
- Panels contain many parameters and interactive controls for setting up input.
- ChimeraX button runs the Phenix command as a separate process and reads results (typically a few minutes).
- New panels display results and assist evaluating newly modeled structure.
- Details of user interfaces in following slides.
Water Placement User Interface
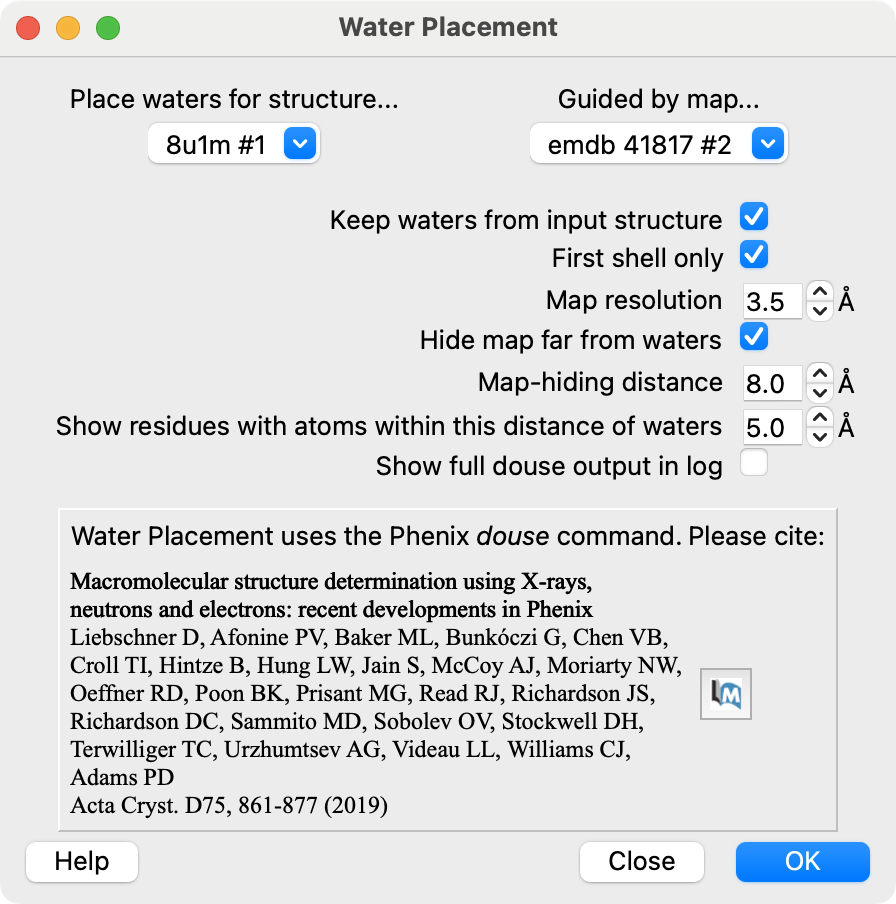
Input model, map and parameters
|
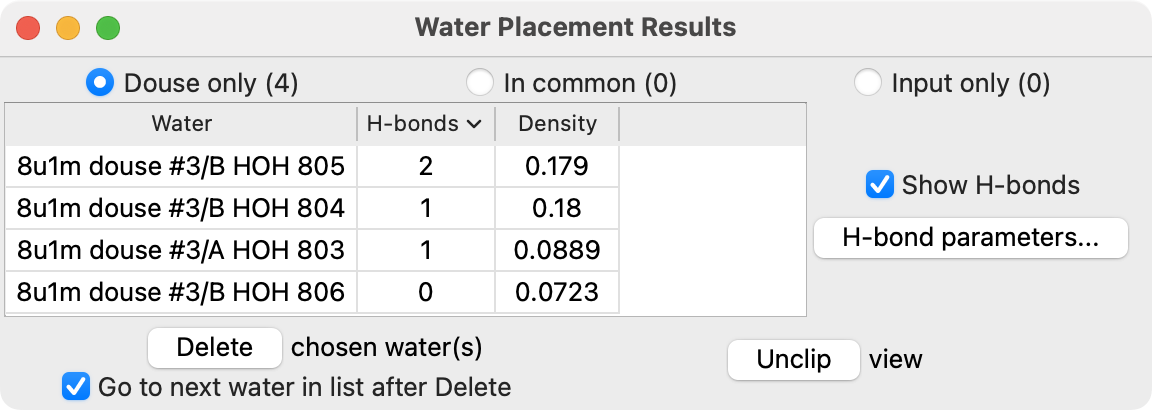
Output table of waters including number of hydrogen bonds
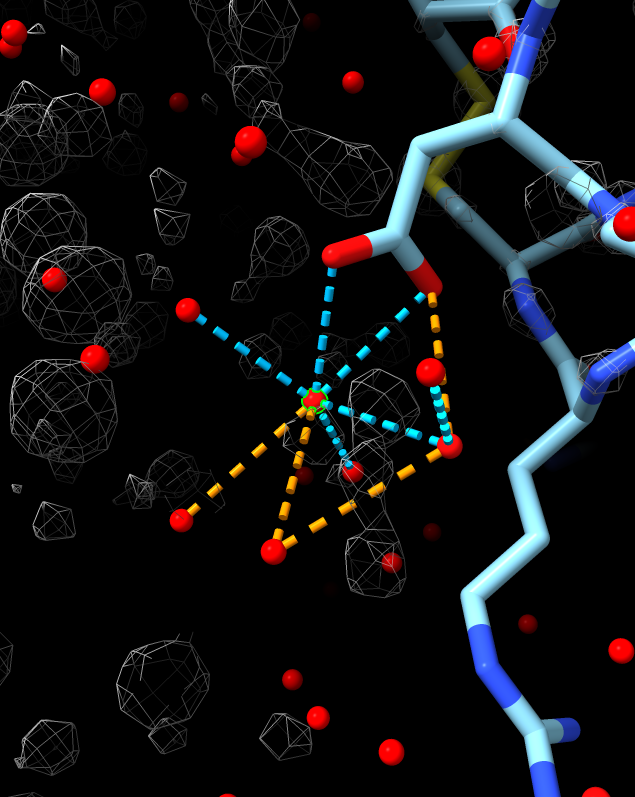
Visualization of water selected in table
|
- The hydrogen bond dashed lines shown are computed by ChimeraX.
- The count is shown in the output table.
- This leverages ChimeraX hydrogen bond detection to assess water placement results.
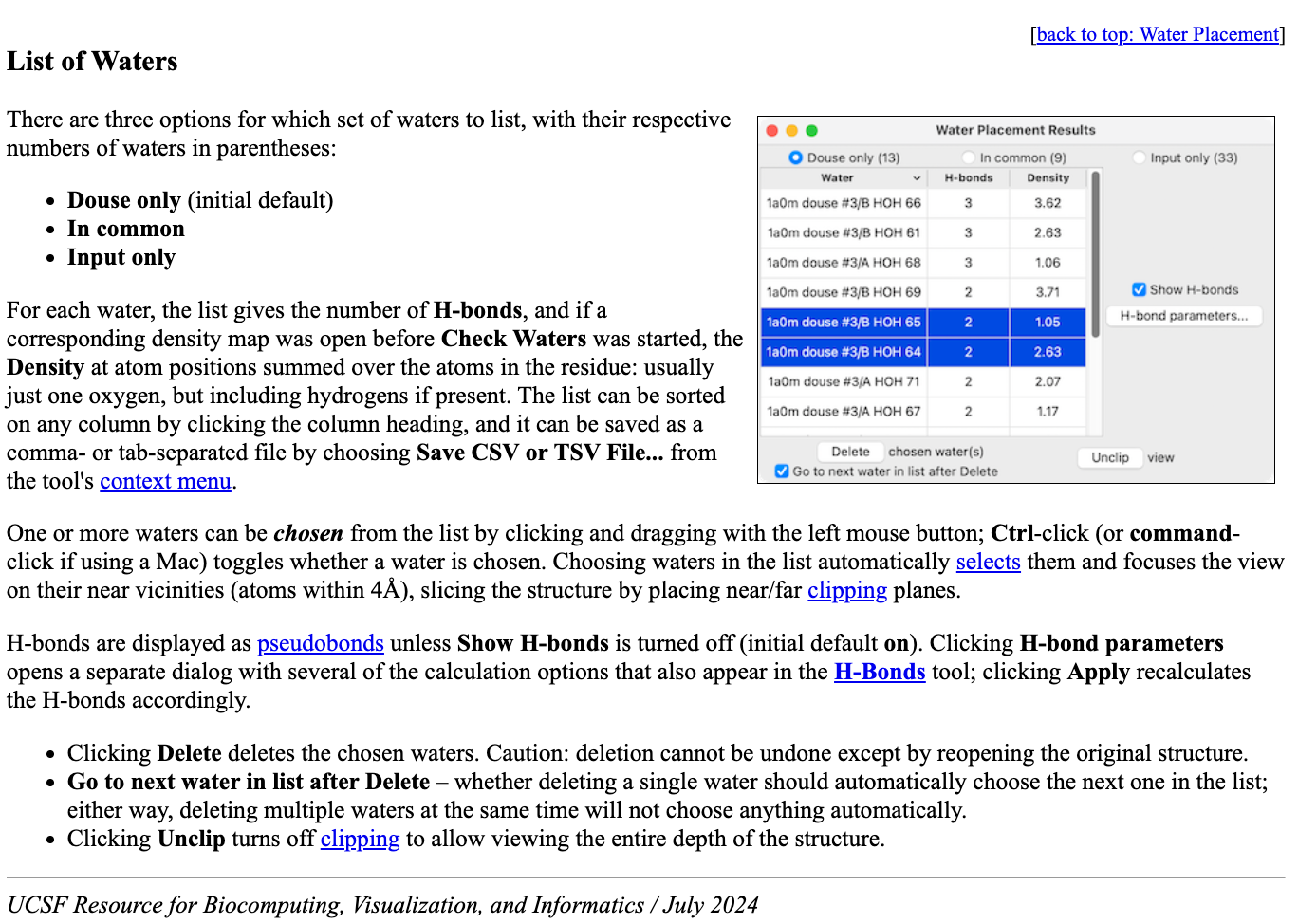
Documentation
All user interfaces have detailed documentation shown in a web browser
by pressing the Help button on the user interface.
All options and mechanisms for exploring the results are
detailed with cross linking to related ChimeraX capabilities.

| 
|
| Water placement documentation
|
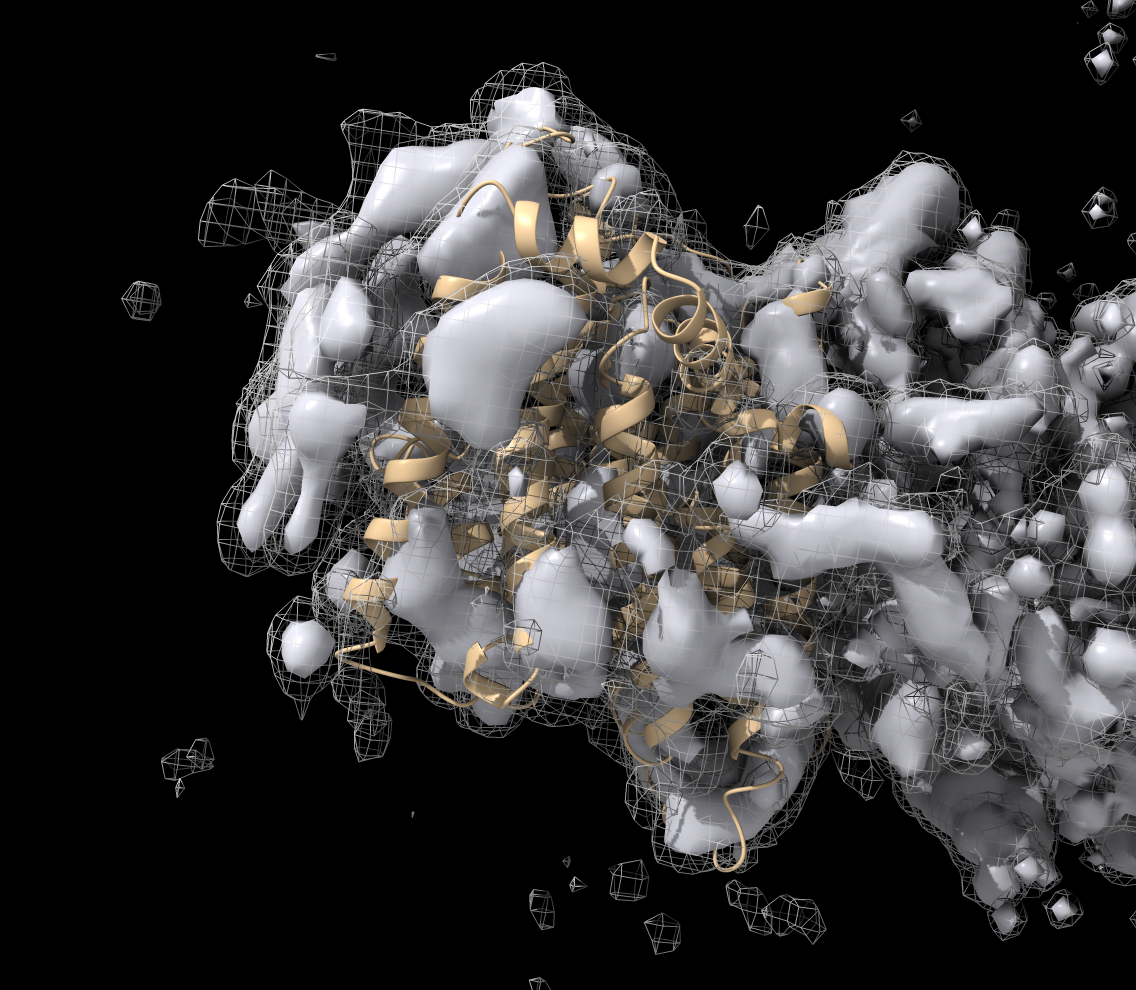
Local EM Fitting User Interface
Video tutorial example made by Dorothee Liebschner (starting at 4:29, about 1 minute).
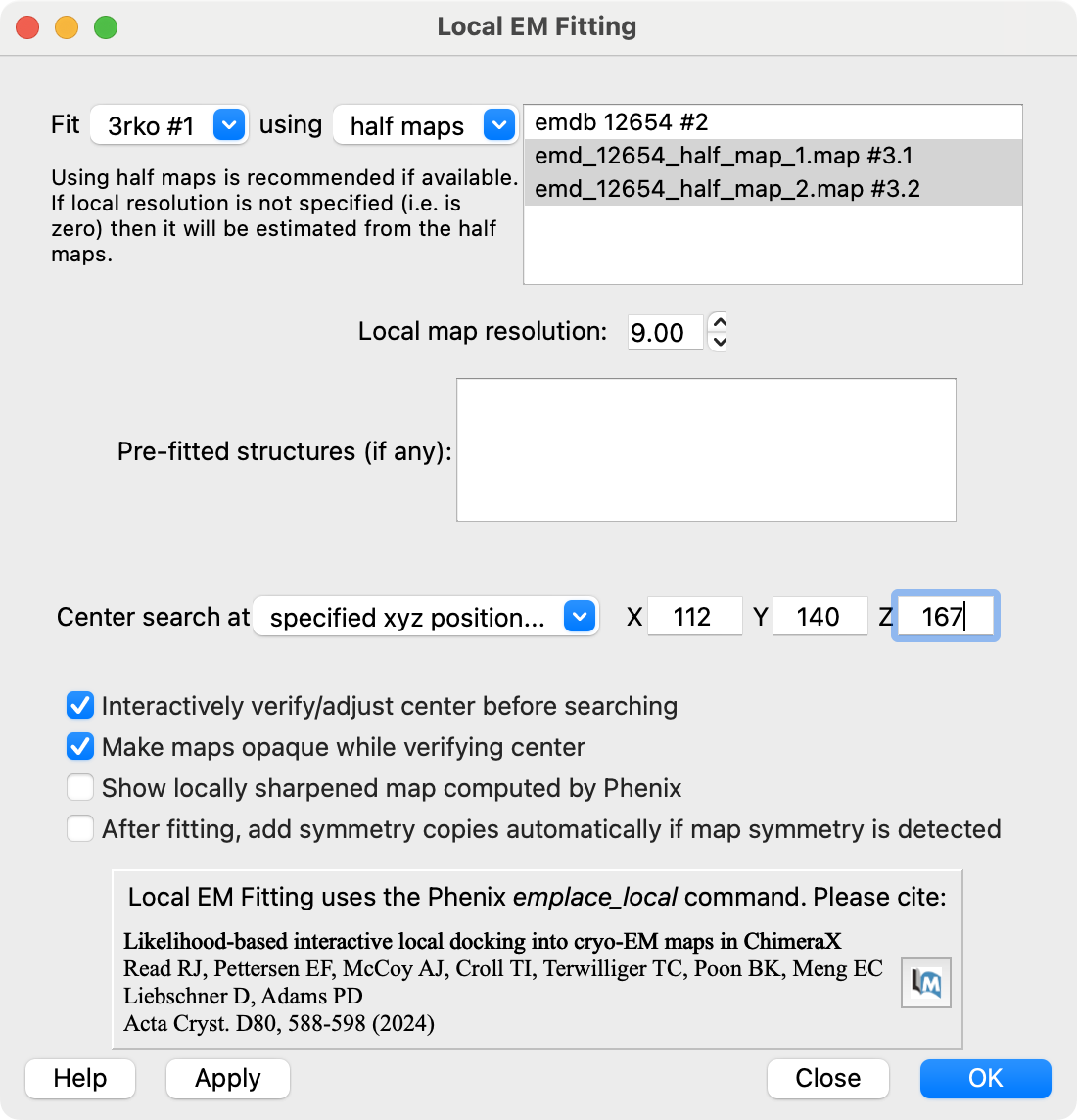
Input model, maps and parameters
|

Orange sphere placed with
mouse to define fitting region.
|
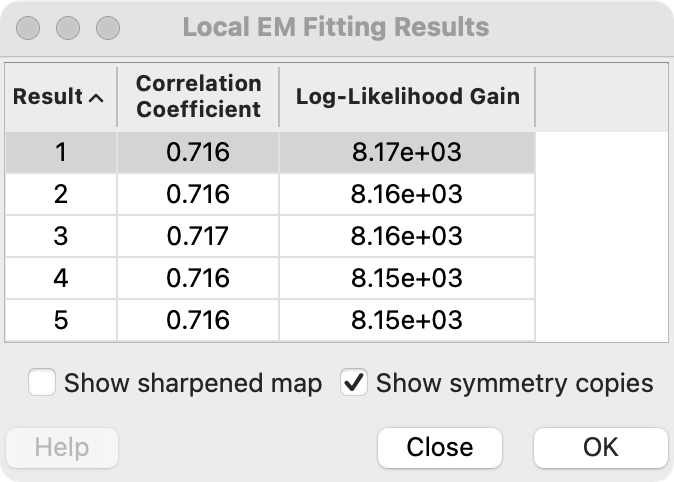
Possible fits shown in a table
Atomic model selected in table
placed at fit position
|
|
Fit Loops User Interface
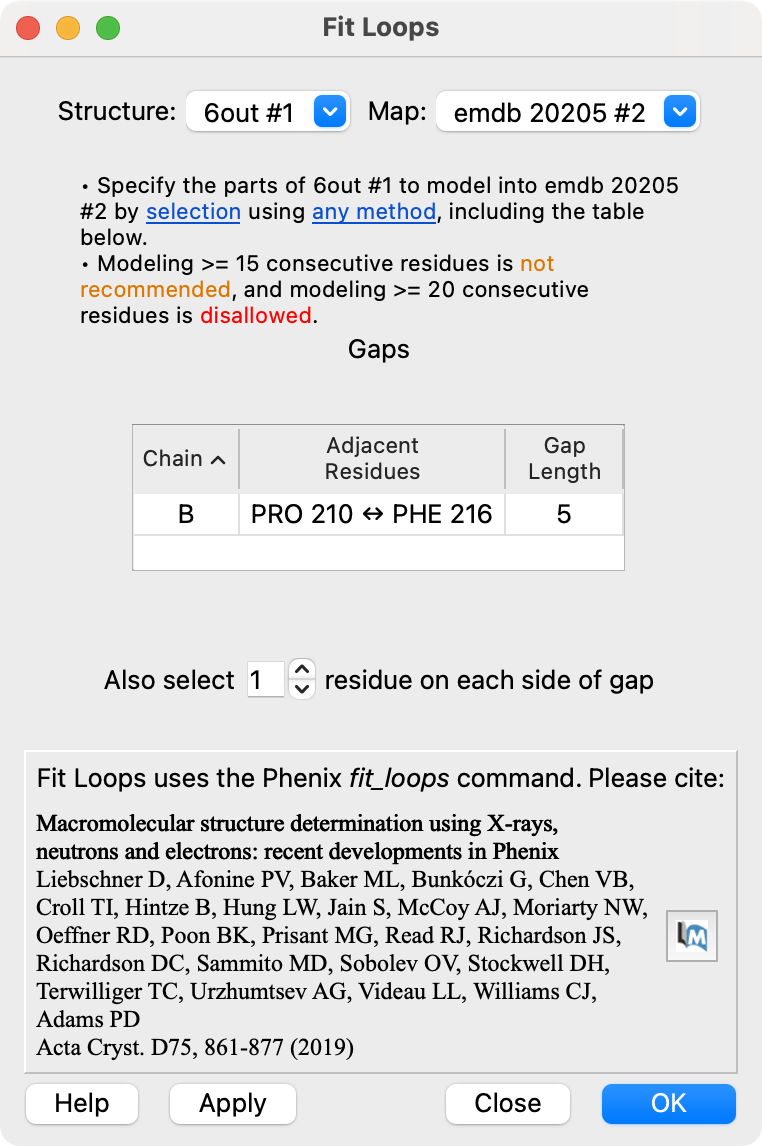
Input model, map and parameters
|
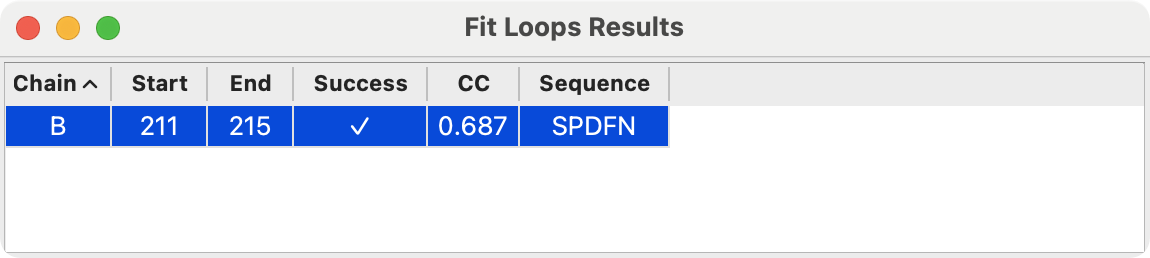
Output table of one or more built loops
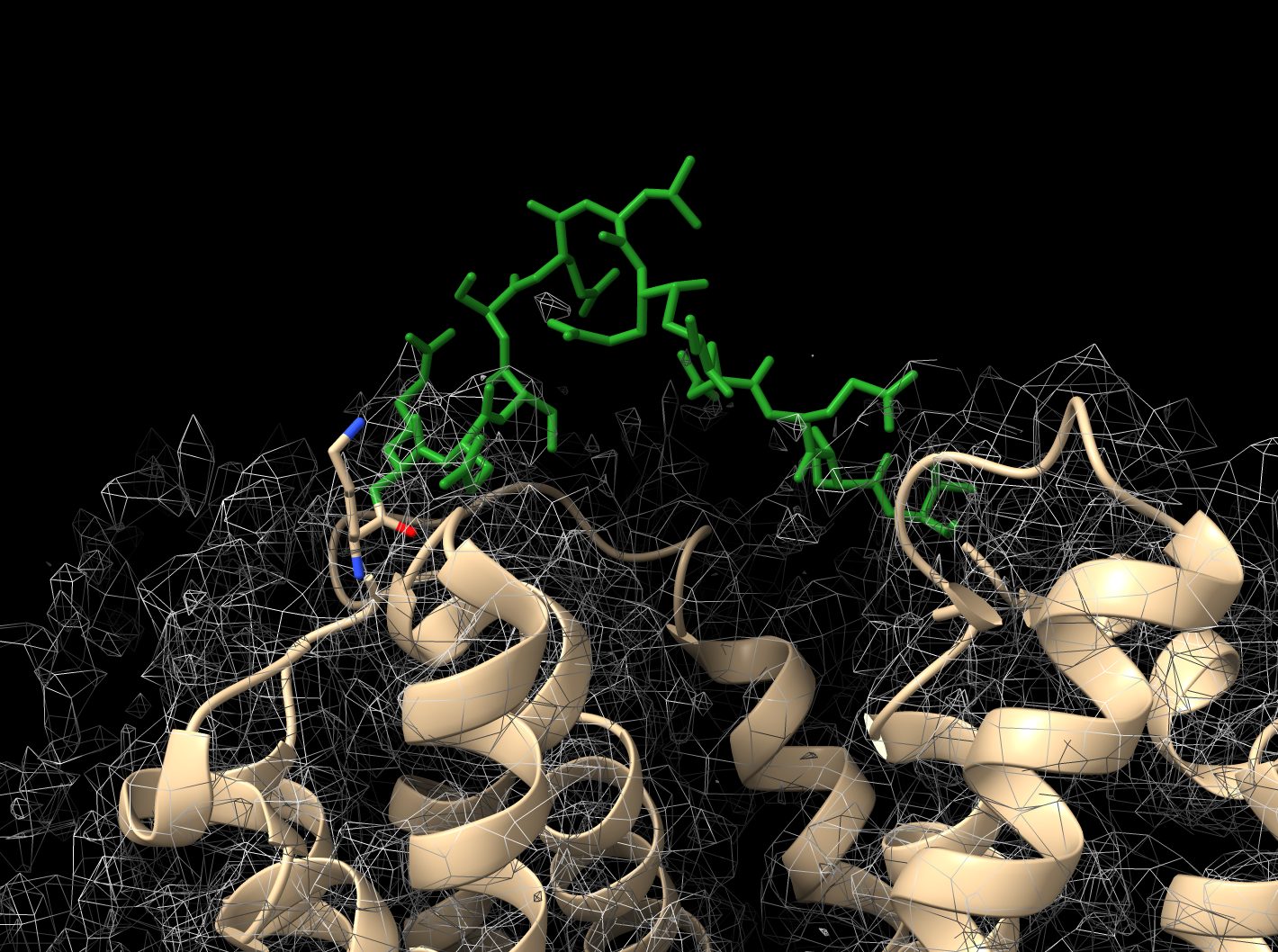
Visualization of new loop (green)
|
Fit Ligand User Interface
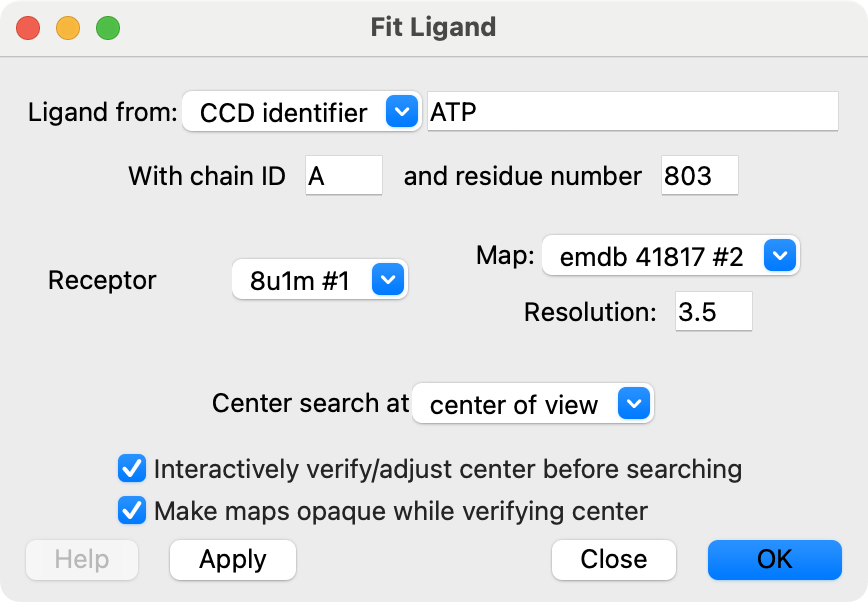
Input model, map and parameters
|
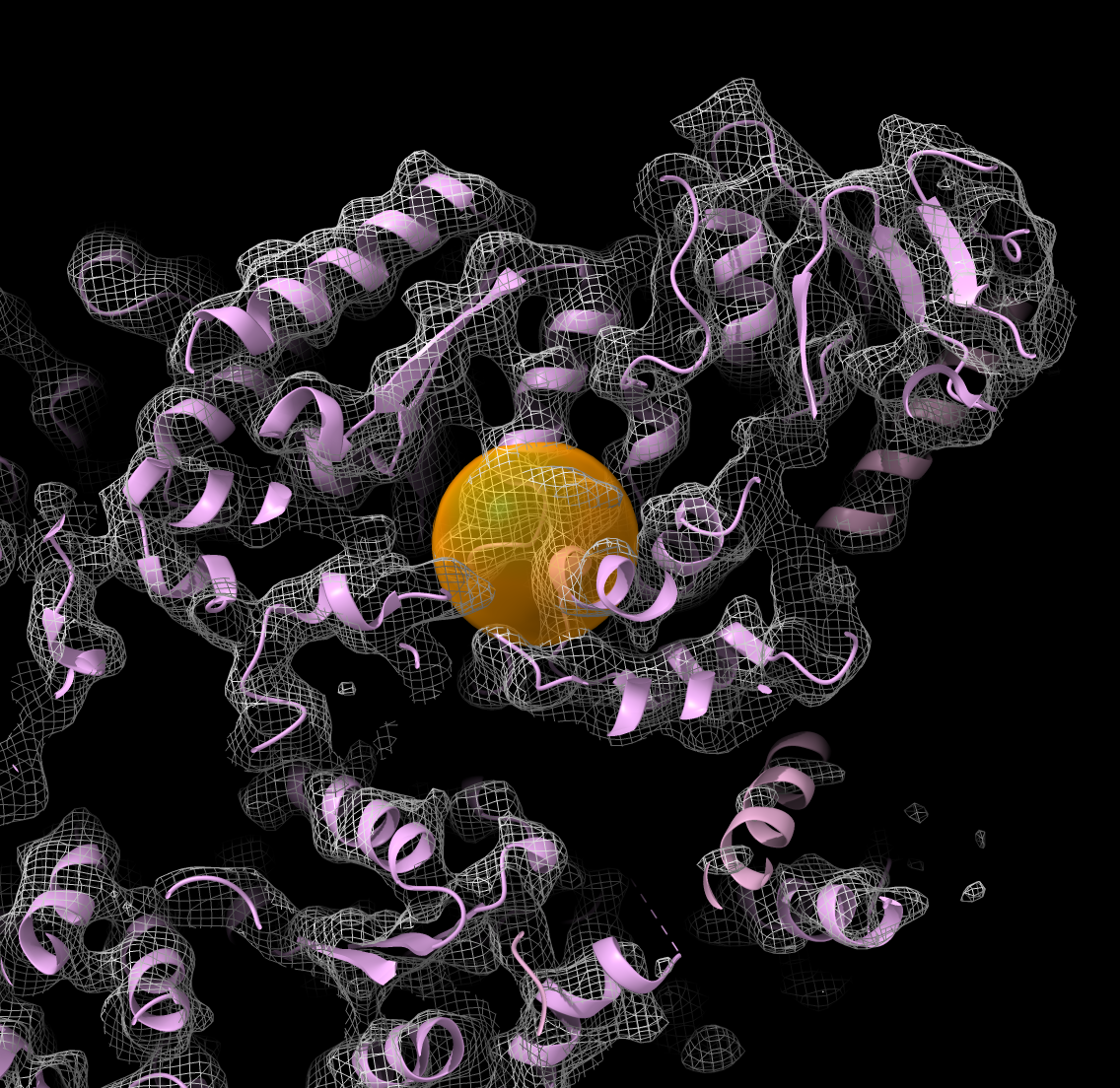
Orange sphere placed with mouse
defines fitting region
|
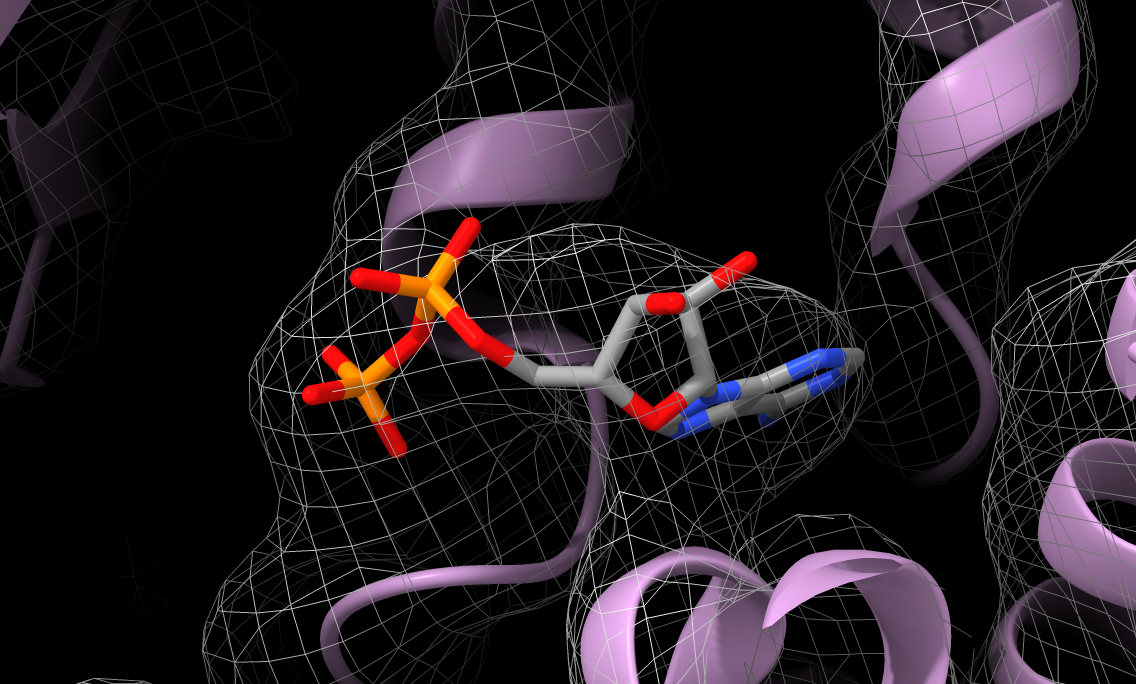
Visualization of ligand fit in density
|
Additional Proposed User Interfaces
The current Phenix R24 proposes two additional user interfaces. We will probably develop one of them
before the current grant ends.
- Restraint specification. Allows a user to specify atomic model building constraints in ChimeraX that are then used by Phenix model building algorithms.
For example:
- the density and model in this region are great - restrain them to their starting geometry
- I have a good reference model from elsewhere for this bit - restrain against that
- The density here is awful, and I've basically just rigid-body docked a domain - restrain all atoms in that part to their starting positions
- Validation. Provides a user interface in ChimeraX for researchers to examine and fix atomic model problems identified by Phenix validation tools such as Molprobity.























