 |
Altloc Explorer lists and manages alternate locations, where an atomic structure contains multiple different positions for the same set of atoms. In PDB format, for example, alternate locations are indicated by a character identifier in column 17 of ATOM and HETATM records.
ChimeraX uses only one of the alternate locations at a time for calculations. For each contiguous (through-bond) set of atoms with alternate locations, the location used initially is that with the highest average atomic occupancy, then as a tie-breaker, the lowest average atomic B-factor, and if a tie-breaker is still needed, by alphabetical order of the alternate location ID. The Altloc Explorer allows changing which alternate locations are used. See also: Rotamers, Build Structure, Dock Prep, Check Waters, Renumber Residues, Change Chain IDs, attributes
The Altloc Explorer can be started from the Structure Analysis section of the Tools menu and manipulated like other panels (more...). It is also implemented as the command altlocs.
The option to Show all altlocs allows viewing multiple alternate locations at the same time without changing which ones are used for calculations on the original structure.. A separate submodel is generated for each alternate location. Initially, these altlocs are shown as thin sticks, but they can be selectively hidden/reshown (see altlocs hide and altlocs show) and their styles, colors, etc. changed, just as in any atomic model.
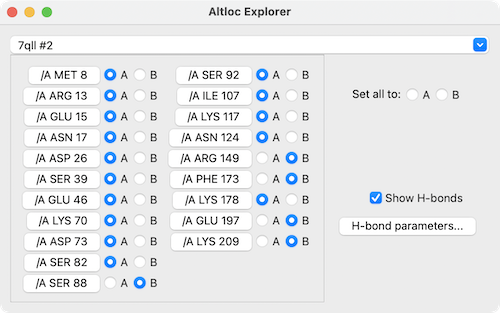
The pulldown menu near the top of the dialog lists the current atomic models. Choosing a specific model lists any residues with alternate locations that it contains. Clicking the identifier of a residue with alternate locations focuses on that residue and displays its atoms, and the adjacent buttons can be used to change which ID (which location) is being used. The Set all to checkboxes allow changing all of the individual residue settings to a specific ID (if available) with a single click.
H-bonds are displayed as pseudobonds unless Show H-bonds is turned off (initial default on). Clicking H-bond parameters opens a separate dialog with several of the calculation options that also appear in the H-Bonds tool; clicking Apply recalculates the H-bonds accordingly.