





A set of tutorials is included in the Chimera User's Guide.
The expanded "Getting Started" tutorial is more suitable for printing (more self-contained rather than hyperlinked) than the above.
Video tutorials and tutorials from past Chimera workshops are also available.
This page contains still more tutorials.
 |
Calculate and visualize APBS electrostatic potential by Thomas Evangelidis. |
 |
Comparing different ligands of the same protein. December 11, 2017. |
 |
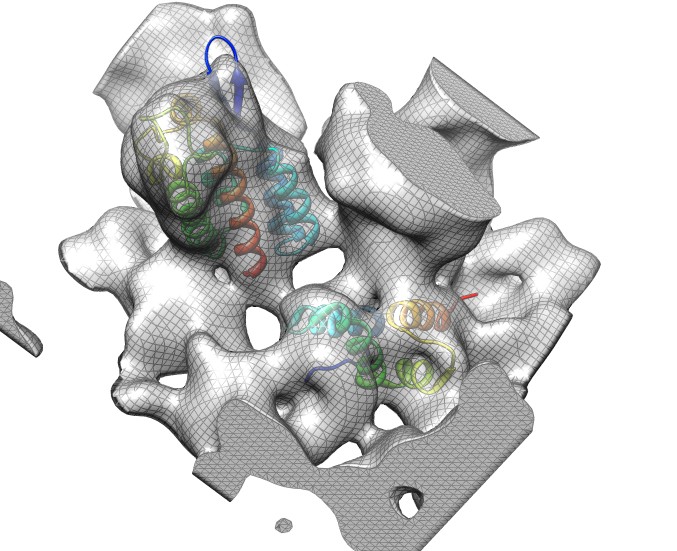
Docking an atomic structure into low-resolution density. December 11, 2017. |
 |
Coloring by NMR chemical shift perturbation. November 29, 2017. Updated version November 28, 2018. |
 |
Mapping sequence conservation onto structures, with several example situations and data files. First presented in Fall 2014. |

| Making animations presented at the NIH Chimera workshop, October 3, 2012. Best viewed in a browser that handles embedded H.264 video (e.g. Chrome or Safari but not Firefox as of Oct 2012). |

| Making high quality images presented at the National University of Singapore cryoEM workshop, July 12, 2012. |

| Filtering and segmenting HIV virus electron tomography presented at the NIH Chimera Workshop, October 2-5, 2012, and at the National University of Singapore cryoEM workshop, July 12, 2012. |
 |
Functional annotation scenario: The Structure-Function Linkage Database (SFLD) and Chimera. 2014 update of a demonstration for an NIH site visit, November 2011. |

| Fitting ParM helical filaments, presented at Purdue University, November 30, 2011. |

| Segger segmentation tutorials from Greg Pintilie at NCMI (2011): Segmenting, fitting molecules to segments, scoring fits. |

| EM fitting tutorial with Rous sarcoma virus and comparison to HIV (data files, video, PDF) presented at the EMAN workshop held in Houston, Texas, March 2011. |

| EM fitting tutorial using HIV spikes and ParM filaments from EMBO practical course "The combination of electron microscopy and x-ray crystallography for the structure determination of large biological complexes" held in Grenoble, France, October 2009. |

| Movie making tutorial from movie making course at UCSF library (May 5, 2009). |

| Movie making tutorial from publication images and movies course (July 29, 2008). |

| Exercises from July 2008 volume data display course. Coloring using electrostatic potential, viewing crystal difference maps, solvent occupancy map, morphing between maps, erasing part of a map, tracing surfaces in tomograms. |

| Density maps. (March 2007). Coloring, morphing, and fitting maps. Plus basic map display and comparison of sequences and structures. Human RNA polymerase II example. |

| Molecular bracelet (February 2007). Creating an image with per-model clipping and silhouette edges; creating a short movie. |

| Chaperonin. Displaying electrostatic potential and crystallographic density map, and fitting model into EM density map. |

| Guide to Volume Data Display in Chimera. Example images and how-to instructions for volume data display features. |

| How to show a virus capsid. Example using Rhinovirus 2 bound to cellular receptor fragment. |

| GroEL tutorial. Density map and PDB model display. |

| Semliki forest virus tutorial. Density map and PDB model display. |