

 about
projects
people
publications
resources
resources
visit us
visit us
search
search
about
projects
people
publications
resources
resources
visit us
visit us
search
search
Quick Links
Featured Citations
In situ structural mechanism of epothilone-B-induced CNS axon regeneration. Bodakuntla S, Taira K et al. Nature. 2025 Dec 11;648(8093):477–487.
Synthetic α-synuclein fibrils replicate in mice causing MSA-like pathology. Burger D, Kashyrina M et al. Nature. 2025 Dec 11;648(8093):409-417.
Multiscale structure of chromatin condensates explains phase separation and material properties. Zhou H, Huertas J et al. Science. 2025 Dec 4;390(6777):eadv6588.
Mechanism of conductance control and neurosteroid binding in NMDA receptors. Kang H, Steigerwald R et al. Nature. 2025 Dec 4;648(8092):220–228.
Membrane-forming phospholipids allosterically modulate native-state prolyl isomerization in a CNG channel. Newton AJ, Latvala RD et al. Protein Sci. 2025 Dec;34(12):e70383.
More citations...News
November 21, 2025
The ChimeraX 1.11 release candidate is available – please try it and report any issues. See the change log for what's new. This will be the last release to support Red Hat Enterprise Linux 8 and its derivatives.
July 24, 2025
ChimeraX 1.10.1 is now available, fixing the problem in 1.10 of repeat registration requests to some users.
June 26, 2025
The ChimeraX 1.10 production release is available! See the change log for what's new.
Previous news...Upcoming Events
UCSF ChimeraX (or simply ChimeraX) is the next-generation molecular visualization program from the Resource for Biocomputing, Visualization, and Informatics (RBVI), following UCSF Chimera. ChimeraX can be downloaded free of charge for academic, government, nonprofit, and personal use. Commercial users, please see ChimeraX commercial licensing.
ChimeraX is developed with support from National Institutes of Health R01-GM129325.
Feature Highlight
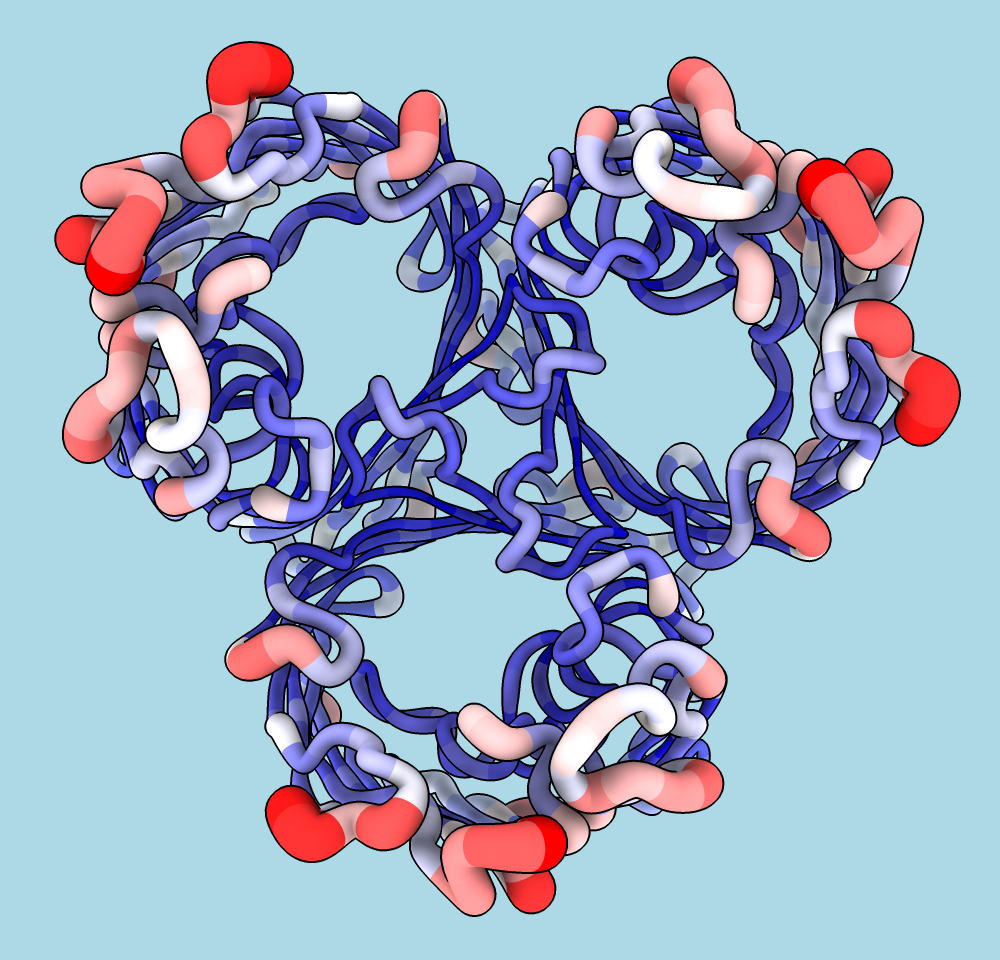
Worms are specialized cartoons in which “fatness”
reflects the values of an
attribute
such as bfactor or seq_conservation.
In the example image, the average atomic B-factor per amino acid residue
is shown with both coloring and worms.
The structure is a trimer of E. coli porin
(PDB 1pho).
For image setup, see the command file
worms.cxc.
Worms are available in ChimeraX v1.8 daily builds 3/12/24 and newer.
Worms and coloring by attribute can be done with the
Render by Attribute
tool or commands
cartoon
byattribute
(aka worm)
and color
byattribute.
Example Image
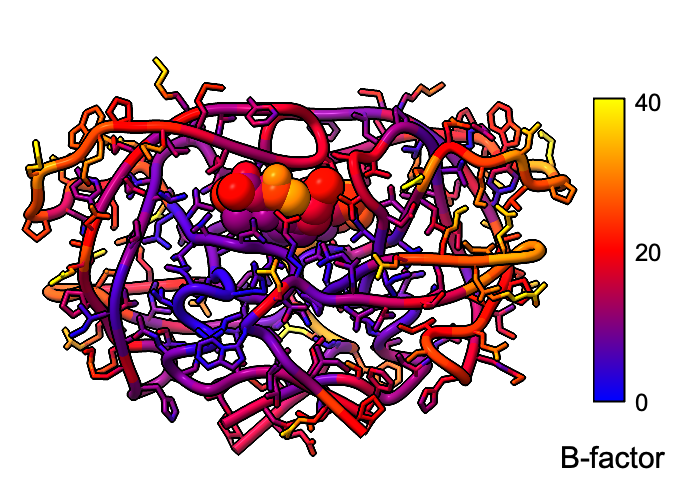
Atomic B-factor values are read from PDB and mmCIF input files
and assigned as attributes
that can be shown with
coloring
and used in
atom specification.
This example shows B-factor variation within a structure of the
HIV-1 protease bound to an inhibitor
(PDB 4hvp).
For complete image setup, including positioning,
color key, and label,
see the command file bfactor.cxc.
Additional color key examples can be found in tutorials:
Coloring by
Electrostatic Potential,
Coloring by Sequence Conservation
Worms
B-factor Coloring
About RBVI | Projects | People | Publications | Resources | Visit Us
Copyright 2018 Regents of the University of California. All rights reserved.